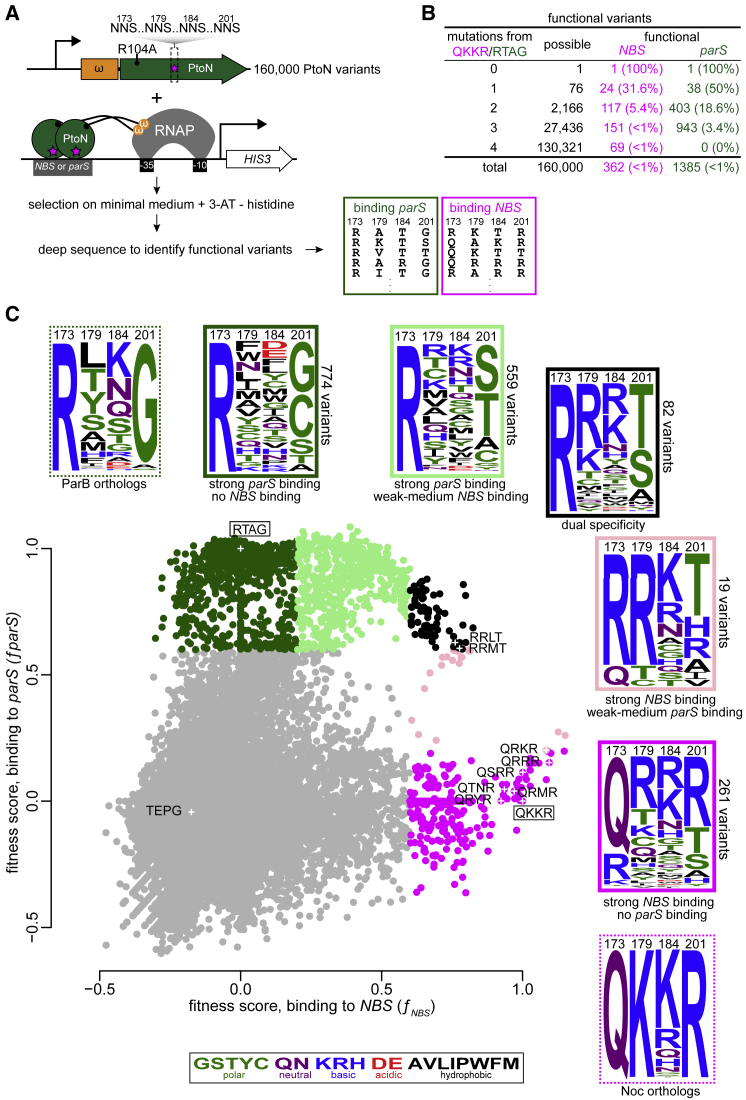
Figure 5.
High-Throughput Mapping of the Fitness of Protein-DNA Interface Mutants
(A) The principle and design of the deep mutational scanning experiment that was based on a bacterial one-hybrid assay and high-throughput sequencing.
(B) Summary of functional NBS-binding and parS-binding variants.
(C) Fitness scores of variants, as assessed by their ability to bind NBS (x axis) or parS (y axis). Dark green: strong parS binding, no NBS binding (fitness score: ƒparS ≥ 0.6, ƒNBS ≤ 0.2); light green: strong parS binding, weak-to-medium NBS binding (ƒparS ≥ 0.6, 0.2 ≤ ƒNBS ≤ 0.6); magenta: strong NBS binding, no parS binding (ƒNBS ≥ 0.6, ƒparS ≤ 0.2); pink: strong NBS binding, weak-to-medium parS binding (ƒNBS ≥ 0.6, 0.2 ≤ ƒparS ≤ 0.6); black: dual specificity (ƒNBS ≥ 0.6, ƒparS ≥ 0.6). Frequency logos of each class of variants are shown together with ones for ParB/Noc orthologs. The amino acids are colored according to their chemical properties. The positions of wild-type (WT) ParB (RTAG), Noc (QKKR), and nine selected variants for an independent validation are also shown and labeled on the scatterplot.