Figure 6.
Deep Mutational Scanning Experiments Reveal the Common Properties of the Mutational Paths to a New DNA-Binding Specificity
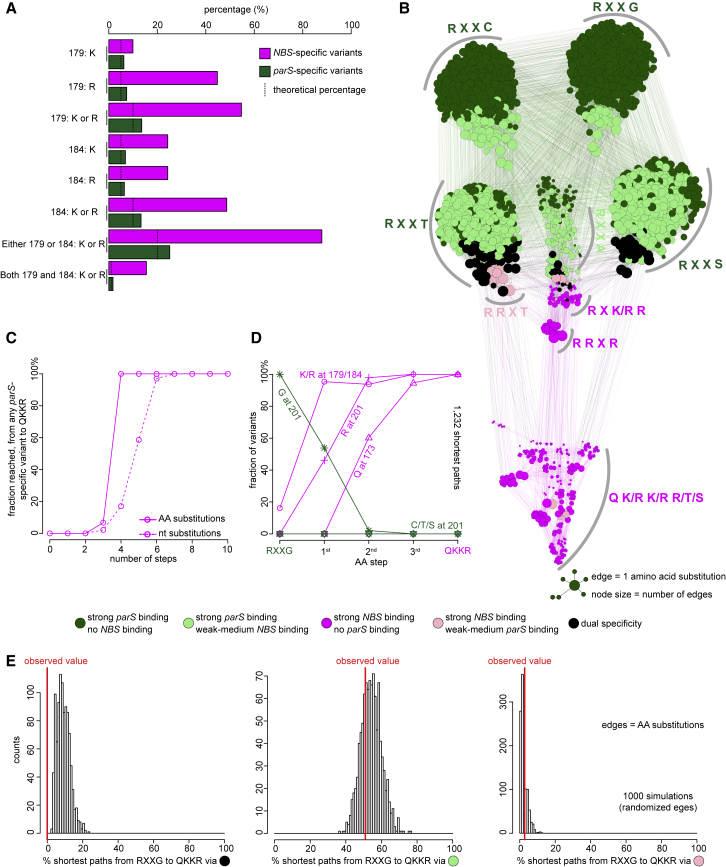
(A) Fractions of arginine or lysine residues at position 179 or 184 or both, in parS-specific (dark green) and NBS-specific (magenta) variants. The dotted lines indicate the expected percentage if arginine/lysine was chosen randomly from 20 amino acids.
(B) A force-directed network graph connecting strong parS-binding variants to strong NBS-binding variants. Nodes represent individual variants, and edges represent single aa substitutions. Node sizes are proportional to their corresponding numbers of edges. Node colors correspond to different classes of variants.
(C) Cumulative fraction of highly parS-specific variants that reached an NBS-specific QKKR variant in a given number of aa (solid line) or nt (dotted line) substitutions (see also Figure S7A).
(D) Fraction of intermediates on all shortest paths from highly parS-specific RXXG variants to the NBS-preferred QKKR that have permissive amino acids (K/R) at either position 179 or 184 or both or have R at position 201, or Q at position 173, or C/T/S at position 201 after a given number of aa steps (see also Figure S7D).
(E) Percentage of shortest paths that traversed black, light green, or pink variants to reach QKKR from any of the highly parS-specific RXXG variants (red lines). The result was compared to ones from 1,000 simulations where the edges were shuffled randomly while keeping the total number of nodes, edges, and graph density constant.