Fig. 4.
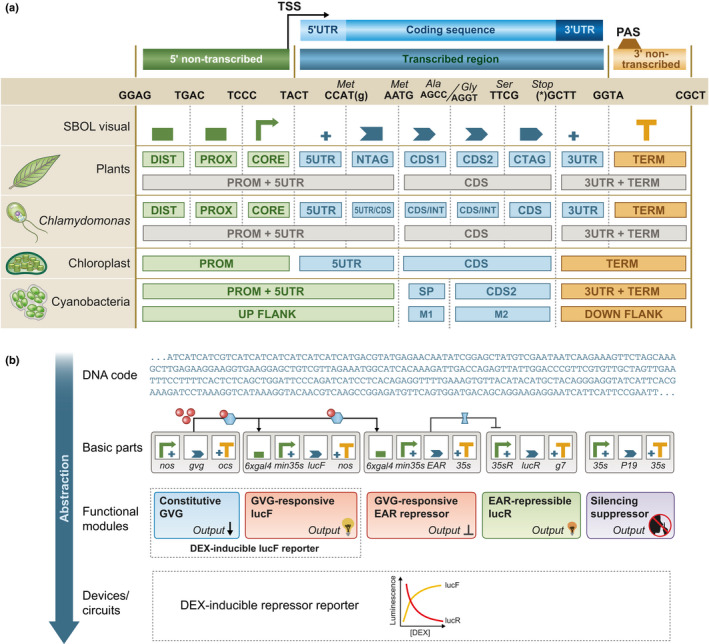
Summary of the Phytobrick standard and its application in abstraction hierarchies. (a) The Phytobrick syntax divides eukaryotic genes into 10 basic functional units: DIST (distal promoter region, cis regulator, or transcriptional enhancer); PROX (proximal promoter region or transcriptional enhancer); CORE (minimal promoter region, including transcription start site); 5UTR (5′ untranslated region); NTAG (amino‐terminal coding region); CDS1 and 2 (coding regions); CTAG (carboxy‐terminal coding region); 3UTR (3′ untranslated region); TERM (transcription terminator, including polyadenylation signal). Each part is represented by a Synthetic Biology Open Language visual (SBOLv) glyph. Phytobrick parts can comprise the region between an adjacent pair of fusion sites or span many sites. Each Phytobrick consists of portion(s) of a gene cloned into a plasmid flanked by a convergent pair of BsaI Type IIS restriction endonuclease recognition sequences. Adaptations for other systems include the assignment of specific part types for use as flanking regions (up‐flank and down‐flank) to enable homologous recombination into the host genome, SP (signal peptide), and as selectable markers (M1 and M2). (b) A synthetic genetic circuit to measure simultaneous repression and activation in response to an external signal (dexamethasone; dex) can be described at four levels of abstraction: DNA level; graphical description of 17 Phytobricks; functional modules of five transcriptional units; a complete circuit for which activity could be quantitatively described.
