Figure 2.
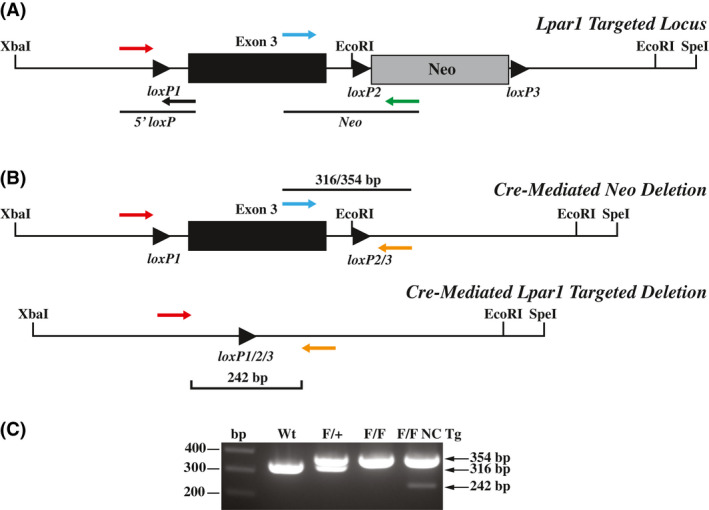
Cre‐mediated deletion in mice with a recombined Lpar1 allele. A, Schematic showing PCR primer pairs used to screen for cre‐mediated deletion of the floxed neomycin cassette. The primers shown assay for the presence of the 5′ loxP site and the presence or absence of the neomycin cassette. B, Diagrams showing the finished Lpar1 targeted allele produced through in vivo cre‐mediated deletion of the neomycin cassette (top) and cre‐mediated targeted deletion of floxed Lpar1 exon 3 (bottom). The three‐primer combination used for PCR genotyping is indicated. C, PCR genotyping of tail DNA from wildtype (Wt), Lpar1flox/+(F/+), Lpar1flox/flox(F/F), and Lpar1flox/flox‐nestin‐cre transgenic (F/F NC Tg) mice. Primers shown in (B) were used for PCR. Wt Lpar1 produced bands of 316 bp, while floxed alleles produced bands 354 bp. The presence of a 242 bp band in the Lpar1flox/flox‐nestin‐cre sample is indicative of cre‐mediated deletion in neural tissue present in the mouse tail.
