Fig. 5.
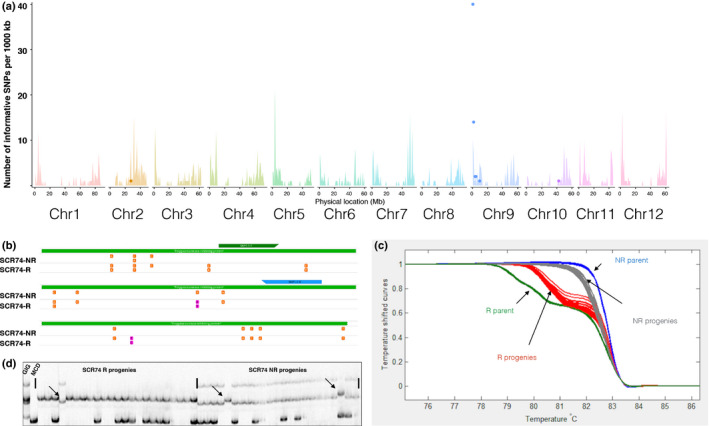
The gene conferring response to SCR74 was mapped to chromosome 9 by receptor‐like protein/kinase enrichment sequencing (RLP/KSeq). (a) Mapping of SCR74. The x‐axis represent the physical positions of the 12 individual potato chromosomes and the y‐axis the number of RLP/receptor‐like kinases (RLKs) or single nucleotide polymorphisms (SNPs) per 1 Mb bin. The background colour spikes represent the number and position of annotated RLP/RLKs genes and the coloured dots depict the position of significant and linked SNPs to SCR74 in a 1 Mb bin. The significant accumulation of SNPs on the top of chromosome 9 indicate the map position of SCR74 receptor. (b) Within the identified region on chromosome 9, a polygalacturonase‐inhibiting protein (PGIP, PGSC0003DMG400006492, green bar) contains 15 SNPs (orange). Two of these SNPs (pink) show a near 1 : 1 frequency in the SCR74‐B3b responsive pool and, together with marker RLP/KSeq‐snp1.1 (green and blue arrow), flank the interval. (c) Melting curves of the high‐resolution melting (HRM) marker RLP/KSeq‐snp1.1 tested on the mapping parents and progenies. (d) Single sequence repeat (SSR) marker STM1051 on chromosome 9 is linked with the SCR74 response. The mapping parents Solanum microdontum ssp. gigantophyllum GIG362‐6 and S. microdontum MCD360‐1, as well as the responsive progenies and nonresponsive progenies were tested with STM1051 and three recombination events (arrow) were found. This figure is reproduced from Domazakis et al. (2017).
