Figure 1.
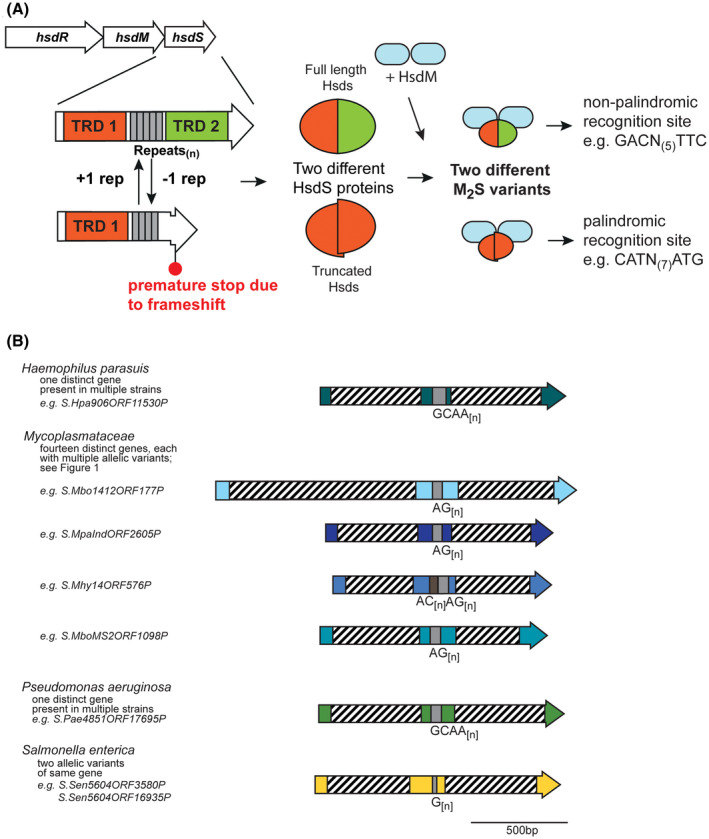
A, Illustration of how phase‐variable switching of hsdS genes occurs. Type I R‐M loci are made up of three genes, encoding a restriction enzyme (hsdR), a methyltransferase (hsdM), and a target sequence specificity protein (hsdS). Each hsdS gene is made up of two target recognition domains (TRDs; TRD 1 in red, and TRD 2 in green), with the SSR tract located between the two TRDs (gray boxes). Loss or gain of repeat units in the SSR tract results in a full‐length hsdS gene being expressed (TRD 1 + 2), which produces a full‐length HsdS protein encoded in a single polypeptide (red/green oval), or a frameshift mutation downstream of the SSR tract, premature transcriptional termination, and results in a truncated HsdS polypeptide (TRD 1 only; red half‐oval). These likely dimerise via the C‐terminal coiled coil region in each truncated HsdS subunit to form a functional HsdS protein. Following oligomerization with an HsdM dimer to form an active methyltransferase, the different HsdS protein subunits result in two different methyltransferase specificities. B, schematic representation of the location of TRD 1 and TRD 2, and the SSR tracts, in a selection of hsdS loci. Colored arrows represent different genes, with color representing homology within each gene if more than one example of this gene is present in REBASE. Hatched boxes represent the locations of the each TRD. The number of different hsdS genes is noted below each species. Unique examples are listed below each individual bacterial species where this hsdS gene is present
