Figure 4.
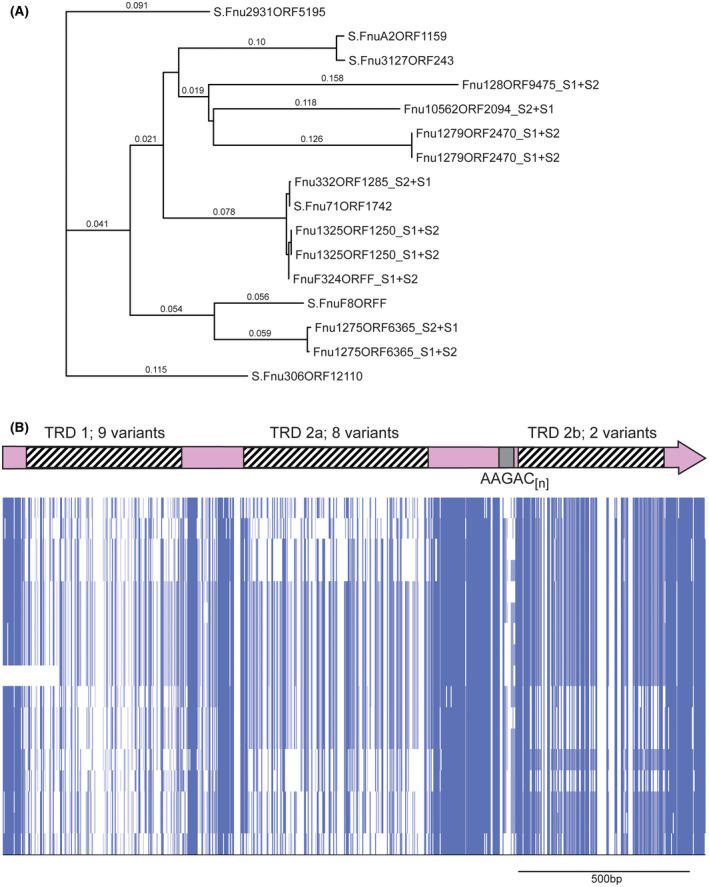
hsdS genes containing AAGAC[n] tracts in Fusobacterium nucleatum. A, a phylogenetic tree was produced by aligning sequences using Muscle, and phylogeny analyzed by RAxML. Where the individual gene is annotated with an “S” prefix, this gene contains an AAGAC[n] repeat tract length where the S1 and S2 regions are in frame, encode the extended HsdS polypeptide, and annotated as a full‐length hsdS gene in REBASE. Where the annotation contains an “S1 + S2” suffix, the AAGAC[n] repeat tract length means that the TRD at the 3′ end of the gene (annotated as TRD 2b here) is out of frame with the 5′ end of the gene encoding TRD 1 and TRD 2a, and annotated as two separate truncated hsdS genes (S1 made up of TRD 1 + TRD 2a, and S2 made up of TRD 2b) in REBASE; B, alignments of the entire hsdS region present in REBASE showing this variation is due to the presence of multiple allelic variants of each of the three TRDs. Sequences were aligned in Muscle, and viewed using JalView overview feature
