Figure 6.
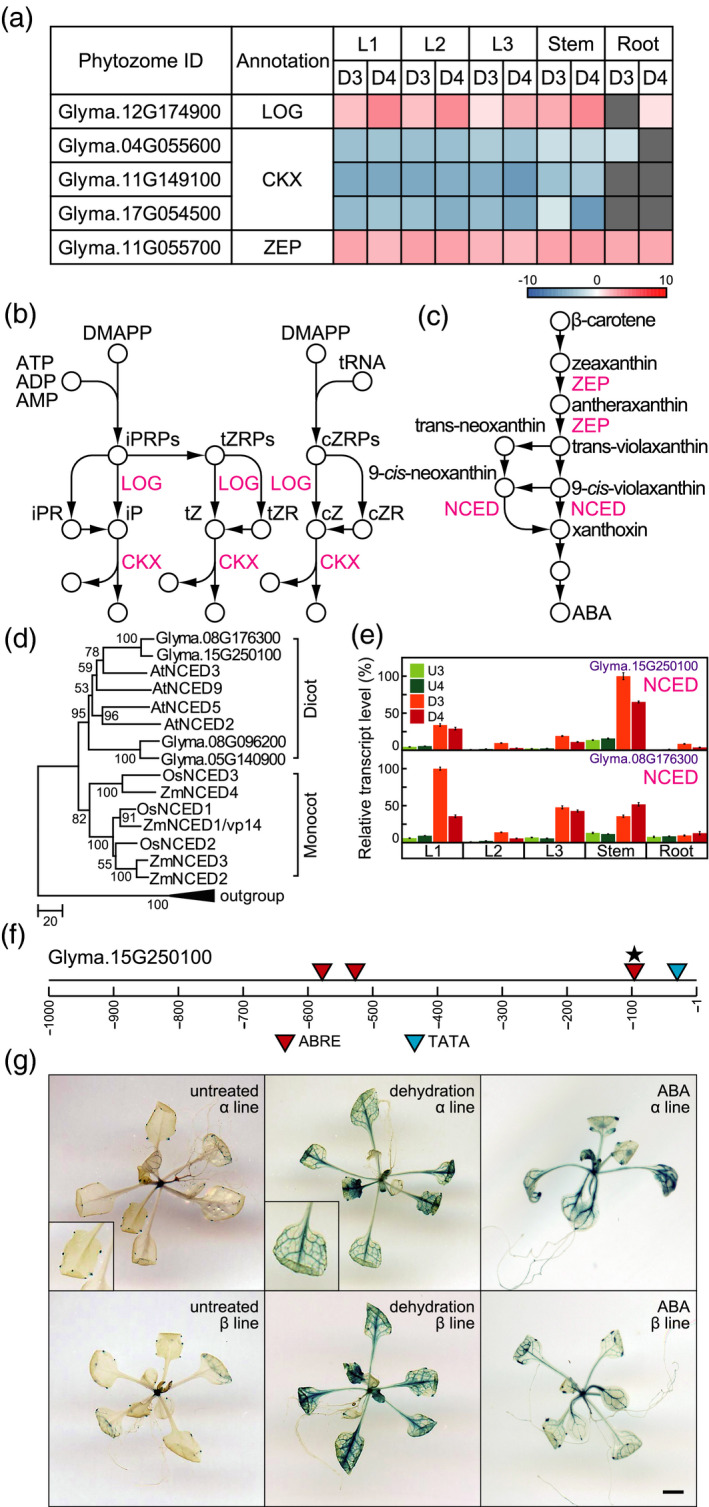
Pathways for CK and ABA biosynthesis and metabolism. (a) Heatmaps illustrate the log ratios of genes involved in cZ and ABA biosynthesis and metabolism. (b) CK biosynthesis and metabolism. (c) ABA biosynthesis. LOG, cytokinin nucleoside 5'‐monophosphate phosphoribohydrolase; CKX, cytokinin oxidase/dehydrogenase; ZEP, zeaxanthin epoxidase; NCED, 9‐cis‐epoxycarotenoid dioxygenase. (d) Phylogenetic trees of NCEDs. The phylogenetic tree was constructed using the neighbour‐joining method with MEGA6 software (Tamura et al., 2013). At3g63520, Glyma.13g202200, GRMZM2G057243, and Os12g0640600 encode carotenoid cleavage dioxygenases and were used as an outgroup. (e) Transcript levels of genes encoding NCED, as determined by RT‐qPCR. Values are shown as the mean ± SD (n = 3 experiments). (f) Schematic diagram of promoters of Glyma.15g250100. Stars indicate duplication of ABREs. (g) Glyma.15g250100 promoter−driven GUS expression in dehydration‐ and ABA‐treated Arabidopsis plants. Left, centre, and right panels show untreated, dehydration‐treated, and ABA‐treated Arabidopsis plants, respectively. The upper and lower panels show two independently isolated lines, α and β. Bar = 0.5 mm.
