Figure 4.
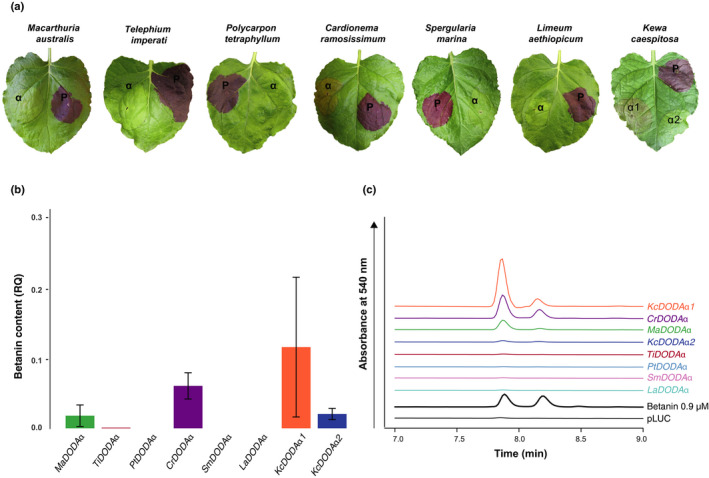
l‐DOPA 4,5‐dioxygenase activity is marginal or absent in DODA from anthocyanin‐producing taxa when heterologously expressed in Nicotiana benthamiana. (a) A representative leaf is shown from the agroinfiltration of l‐DOPA 4,5‐dioxygenase (DODA) genes from Macarthuria australis (MaDODA α), Telephium imperati (TiDODA α), Polycarpon tetraphyllum (PtDODA α), Cardionema ramosissimum (CrDODA α), Spergularia marina (SmDODA α), Limeum aethiopicum (LaDODA α) and Kewa caespitosa (KcDODA α 1, KcDODA α 2). The DODA coding sequences were cloned into multigene constructs with the other structural genes necessary for betacyanin production (BvCYP76AD1, MjcDOPA‐5GT). The pBC‐BvDODAα1 multigene vector was included as a positive control (P). (b) Betanin content of the infiltration spots was measured using HPLC. Amounts of betanin were calculated relative (RQ) to the average amount of betanin present in BvDODAα1 infiltration spots for each species and the data combined into one graph (see Supporting Information Fig. S12 for species‐specific data). Bars show means ± SD; n ≥ 3. (c) A representative HPLC trace for each DODA α gene from each species is shown. The left peak is betanin and the right peak is its isomer, isobetanin. The traces are offset for presentation. ‘pLUC’ is a negative control plasmid carrying the firefly luciferase gene and ‘Betanin’ is a commercially available extract from beet hypocotyl, which was used to validate the retention time of betanin in these samples.
