Figure 6.
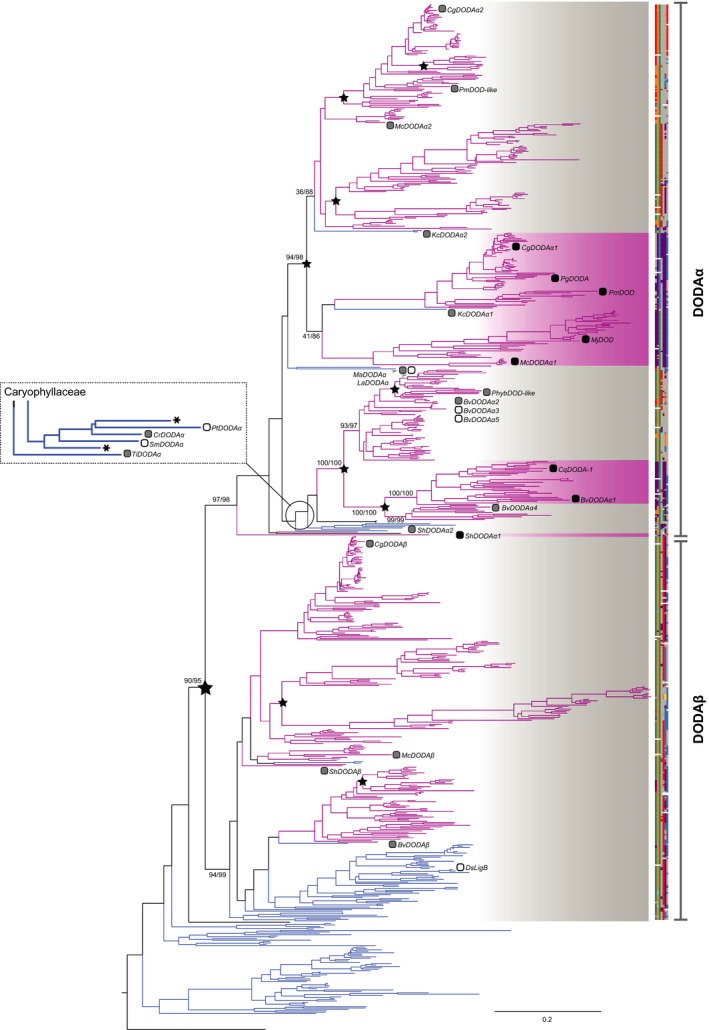
The DODA gene tree shows a homoplasious distribution of functionally characterised DODA genes that produce a high level of betalain pigments. The maximum likelihood phylogeny of Caryophyllales l‐DOPA 4,5‐dioxygenase (DODA) genes was inferred from coding sequences derived from genomes and transcriptomes. Branch lengths are expected number of substitutions per site. Scale bar gives 0.2 expected substitutions per site. Branch labels are support values for major paralogous clades from rapid bootstrapping and the SH‐like Approximate Likelihood Ratio Test, respectively, given as RBS/SH‐aLRT. Putative major duplication nodes are highlighted with stars. Branches are coloured according to the putative pigmentation state of the taxa (blue, anthocyanin; pink, betalain). Labelled tips show functionally characterised DODAs and shaded squares correspond to DODA activity (white, no activity; grey, marginal activity; black, high activity). Asterisks indicate putative pseudogenes. Annotated at tips is a colour‐coded alignment of the seven residues conferring high activity reported in Bean et al. (2018). XxDODA: Bv, Beta vulgaris; Cg, Carnegiea gigantea; Cr, Cardionema ramosissimum; Ds, Dianthus superbus; Kc, Kewa caespitosa; La, Limeum aethiopicum; Mc, Mesembryanthemum crystallinum; Ma, Macarthuria australis; Mj, Mirabilis jalapa; Pg, Portulaca grandiflora; Phyb, Ptilotus hybrid; Pm, Parakeelya mirabilis; Pt, Polycarpon tetraphyllum; Sh, Stegnosperma halimifolium; Sm, Spergularia marina; Ti, Telephium imperati.
