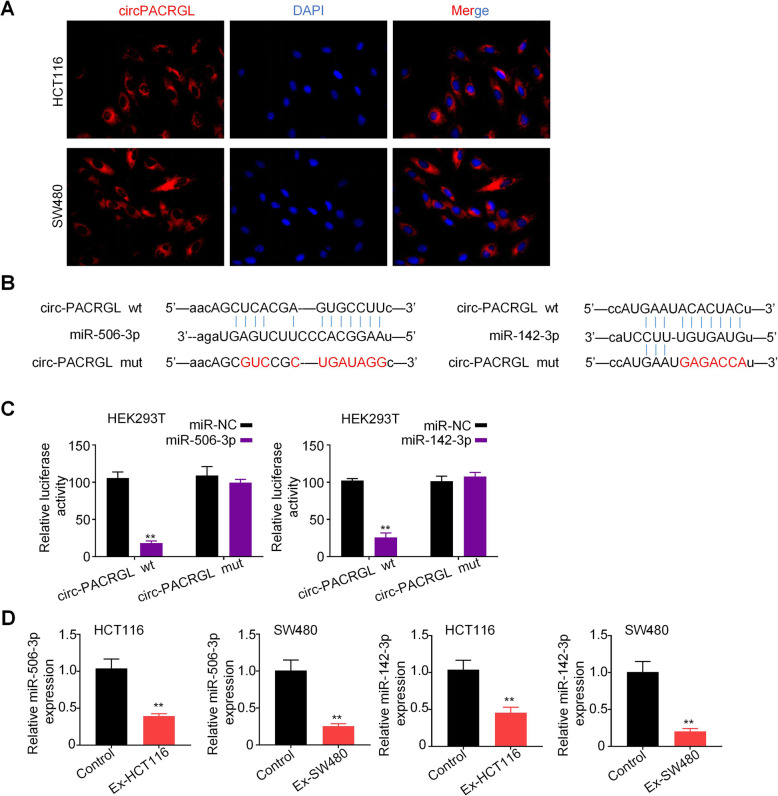
Fig. 3.
circPACRGL serves as a sponge for miR-142-3p and miR-506-3p. a HCT116 and SW480 cells were stained with circPACRGL probe (red) and a nuclear marker (DAPI, blue), and imaged under confocal microscopy after treatment with exosomes (1800× magnification). b Schematic representation of the 3′-UTR of circPACRGL with the predicted target site for miR-506-3p and miR-142-3p. The mutant site of circPACRGL 3′-UTR is indicated (without line). c Luciferase reporter analysis was performed to examine the binging ability between miR-506-3p/miR-142-3p and circPACRGL. Reporter constructs containing either circPACRGLwt or circPACRGLmut at the predicted miR-142-3p/miR-506-3p target sequences were co-transfected into HEK293T cells, along with miR-142-3p/miR-506-3p or miR-NC mimics. d qRT-PCR analyses of miR-506-3p/miR-142-3p expression in HCT116 and SW480 cells after exosomes treatment (Ex-HCT116 and Ex-SW480). All P values were determined by a two-tailed unpaired student’s t-test (**, P < 0.01); wt, wild type; mut, mutation. Ex, exosomes