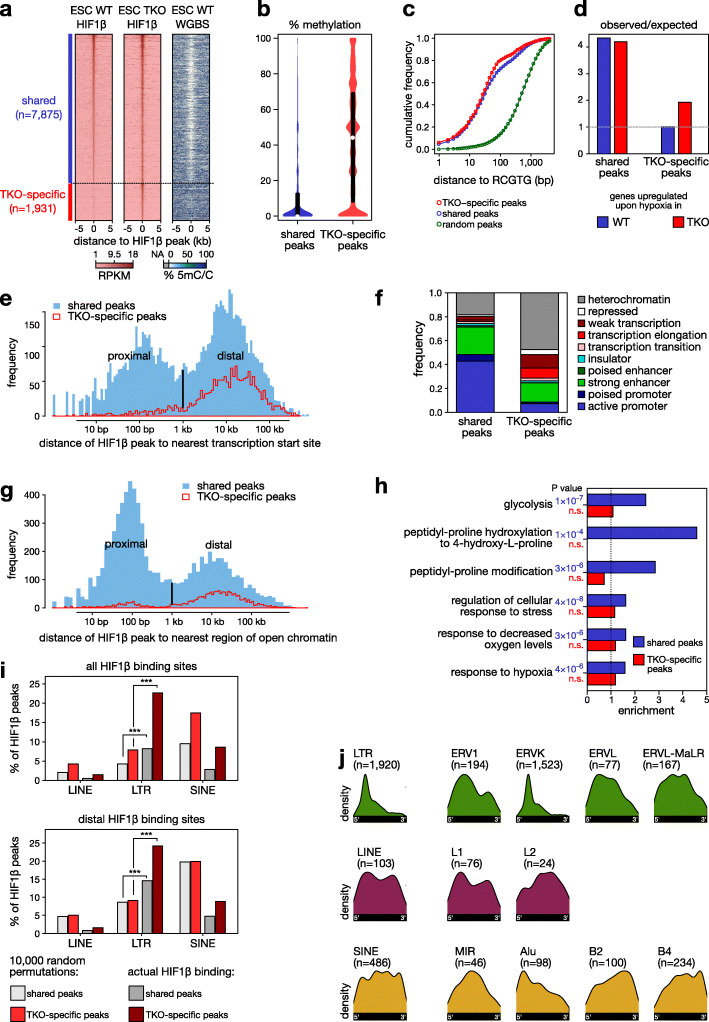
Fig. 3.
DNA demethylation uncovers new HIF1β binding sites. a Heatmaps of HIF1β binding (RPKM) and DNA methylation as determined using WGBS at regions flanking the summit of HIF1β binding peaks (± 5 kb) either shared with WT or TKO-specific ESCs. b % methylation at shared and TKO-specific HIF1β binding sites in WT ESCs. See Additional file 1: Fig. S5 for scatter plots illustrating the correlations between HIF1β ChIP-seq replicates in Dnmt-WT versus Dnmt-TKO ESCs. c Cumulative frequency of distance to the nearest RCGTG motif for shared, TKO-specific, and randomized HIF1β binding peaks. d Observed/expected frequency of upregulated genes associated with shared and TKO-specific HIF1β binding peaks in WT and Dnmt-TKO ESCs exposed to 24 h of hypoxia (0.5% O2). e Distance of shared and TKO-specific HIF1β binding peaks in ESCs to the nearest TSS. A bimodal peak was detected indicating proximal and distal binding events. f Functional genome annotation using ChromHMM of shared and TKO-specific HIF1β binding peaks in ESCs. g Distance of shared and TKO-specific HIF1β binding peaks to open chromatin regions in ESCs. A bimodal peak was detected indicating proximal and distal binding events. h Ontology analysis of genes associated with shared and TKO-specific HIF1β binding peaks in ESCs. i HIF1β binding sites in LINEs, LTRs, and SINEs after 10,000 random permutations and as observed by HIF1β ChIP-seq (actual HIF1β binding) for all HIF1β sites (top panel) and only for distal HIF1β sites (bottom panel). *** P < 0.001 by Fisher’s exact test. j Distribution of HIF1β binding peaks detected in murine Dnmt-TKO ESCs for the retrotransposon families, color-coded by retrotransposon class (green: LTR; violet: LINE; yellow: SINE)