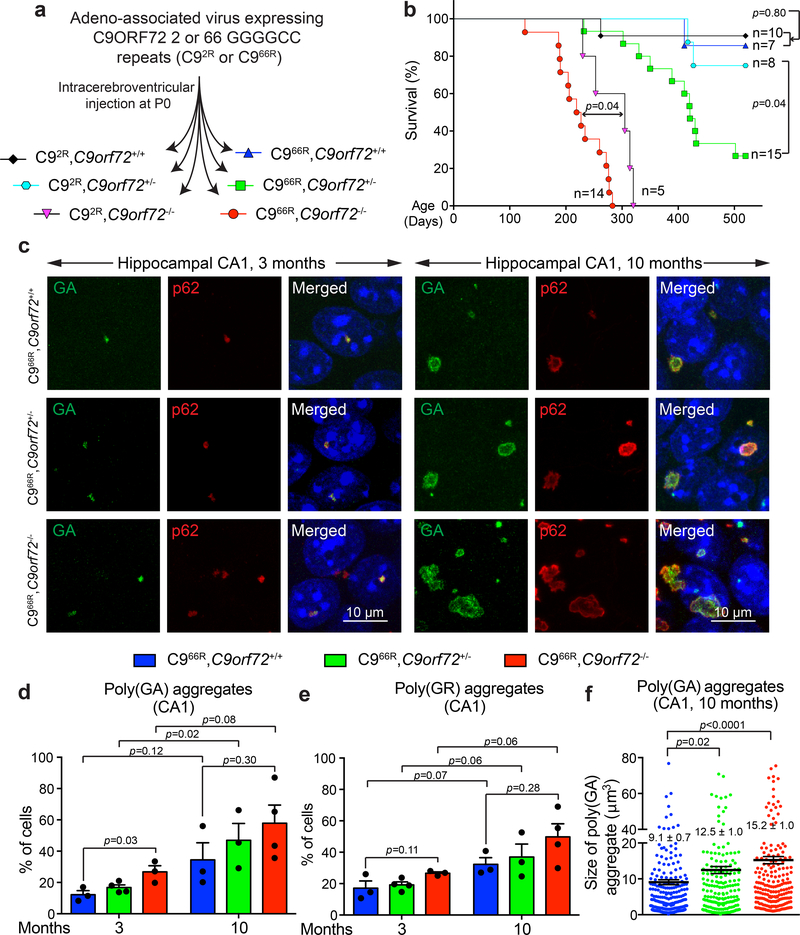
Figure 1: Reduction or loss of C9ORF72 in mice expressing 66 GGGGCC repeats induces or accelerates, respectively, premature death and leads to increased dipeptide-repeat protein accumulation.
(a) Schematic of intracerebroventricular injection of C9AAV in P0 mice to produce cohorts of mice expressing 2 or 66 GGGGCC repeats with neither, one or both endogenous C9orf72 alleles inactivated (C92R,C9orf72+/+; C92R,C9orf72+/−; C92R,C9orf72−/−; C966R,C9orf72+/+; C966R,C9orf72+/−; C966R,C9orf72−/−).
(b) Survival curve (up to 520 days) of the mouse cohorts in (a). Statistical evaluation using the Gehan-Breslow-Wilcoxon test.
(c) Representative images of poly(GA) aggregates or p62 in the hippocampal CA1 region of (left) 3- or (right) 10-month-old mice expressing 66 GGGGCC repeats with normal, reduced, or no endogenous C9ORF72. Experiment was reproduced three times independently with similar results.
(d-e) Percentage of cells with detectable poly(GA) (d) or poly(GR) (e) in the cortex of 3- and 10-month-old mice expressing 66 GGGGCC repeats with normal, reduced or absence of endogenous C9ORF72. Error bars represent SEM (for 3-month-old animals, n = 3 C966R,C9orf72+/+, n = 4 C966R,C9orf72+/−, n = 3 C966R,C9orf72−/−; for 10-month-old animals, n = 3 C966R,C9orf72+/+, n = 3 C966R,C9orf72+/−, n = 4 C966R,C9orf72−/−; over 80 cells counted per animal). Between different groups of genotypes, statistical evaluations were performed using one-way ANOVA with Tukey’s post hoc test. Between different groups of ages within the same genotype, statistical evaluations were performed using student’s t-test, unpaired, two-tailed.
(f) Aggregate size of poly(GA) aggregates in the hippocampal CA1 region of 10-month-old mice expressing 66 GGGGCC repeats with normal, reduced, or absence of endogenous C9ORF72. Each dot represents the size of a poly(GA) aggregate (n = 3 animals per group). Error bars represent SEM. Statistical evaluations were performed using one-way ANOVA with Tukey’s post hoc test.