Summary
Gene regulatory networks reveal how transcription factors contribute to a dynamic cascade of cellular information processing. Recent advances in technologies have enhanced the toolkit for testing GRN mechanisms and connections. Here we emphasize three approaches that we have found important for interrogating transcriptional mechanisms in echinoderms: single cell mRNA sequencing (Drop-seq), nascent RNA detection and identification, and chromatin immunoprecipitation (ChIP). We present these applications in order since it is a logical experimental protocol. With preliminary information from bulk mRNA transcriptome analysis and differential gene expression studies (DE-seq), one may need to test in what specific cells important genes may be expressed and to use single cell sequencing to define such links. Nascent RNA analysis with the Click-iT chemistry allows the investigator to deduce when the RNA was transcribed, not just identify its presence, and ChIP allows the investigator to study direct interactions of putative transcriptional regulators with the gene promoter of interest. This flow of thinking, and the technologies to support it, are presented here for echinoderms. While many of the procedures are general and applicable to many organisms and cell types, we emphasize unique aspects of the protocols for consideration in using echinoderm embryos, larvae, and adult tissues.
Keywords: single cell mRNA-seq, drop-seq, chromatin immunoprecipitation, Click-iT nascent RNA labeling, embryo dissociation
A. Single cell sequencing
1. Transcriptomics and utility of single cell mRNA sequencing
All cells of the body come from a single cell, and the many siblings resulting from cell divisions diversify to acquire a plethora of fates. Understanding the mechanism of this developmental process requires examining each cell apart from its neighbors. Current technology is limited in how we can analyze proteins, lipids, sugars, and metabolites, but polymers of nucleic acids can be replicated experimentally in vitro, making them more abundant and tractable. Investigators recently have been able to do so from a single, isolated cell. Thus, we can now visualize one part of a cell’s fingerprint that drives their form, fate, and function, and begin to understand how diverse a cell type, a tissue, and an organism really is.
This section summarizes the procedure of single cell mRNA sequencing in echinoderms. Now, instead of generating bulk transcriptomes from whole populations of embryos or specific cell types, one can generate transcriptomes from individual cells representing either a specific germ layer, a specific developmental stage, or a tissue type.
The protocol for mRNA-drop sequence analysis is common for many cells from diverse organisms. These protocols have been applied to organs, tissues, and cell types as diverse as human blood, zebrafish embryos, and adult Hydra (Camp, Wollny, & Treutlein, 2018; Fletcher, Das, & Ngai, 2018; Papalexi & Satija, 2018). The controllers for making individual cell/drops are diverse, but all share common principles. The ways in which each population of mRNA from a single cell is processed is also diverse, but again with shared principles. We will summarize these various considerations here for single cell RNA-seq, and emphasize how echinoderms have been processed for drop-seq analysis in order to maximize utility of the technology. Overall, echinoderm embryos, larvae, and adult tissues are wonderfully well suited for such analysis, and the results from these analyses should be transformative over the years to come.
2. Summary of current instrumentation available for single cell sequencing
a. Comparison of Cost, Benefit, Yield
Single-cell RNA sequencing (scRNA-seq) allows mRNA transcriptome wide analysis of individual cells. This involves three major steps; isolating individual cells, extracting RNA and converting it to cDNA, and finally creating a cDNA sequencing library. Several methods are available to perform single cell sequencing analysis. These methods vary in the way they isolate cells and generate cDNA, and as a result they vary in their sensitivity and accuracy (among other things i.e. cost).
A recent review (Ziegenhain et al., 2017) summarizes and analyzes six methods for performing scRNA-seq discussing accuracy, sensitivity, power, and cost efficiency.
CEL-seq2
DROP-seq
MARS-seq
SCRB-seq
Smart-seq
Smart-seq2
All single cell methods require tissue dissociation followed by cell isolation. Cell isolation can be plate based, as with CEL-seq2 and Smart-seq. Cells can be sorted via Fluorescence Activated Cell Sorting (FACS) into a 96-well or 384-well plate, which is the case for MARS-seq, SCRB-seq, and Smart-seq2. In the Drop-seq method, controlled fluid flows interact such that cells are isolated in microdroplets within a microfluidic chamber.
b. Sensitivity and Accuracy
Sensitivity refers to the ability to capture an mRNA transcript for inclusion in the cDNA sequencing library. To address sensitivity, the number of genes detected per cell is measured and this value is compared across the six methods. Smart-seq2 showed the highest sensitivity measure and Drop-seq the lowest.
Accuracy refers to how well the read quantification matches the concentration of mRNA transcripts. To measure accuracy, synthetic RNA transcripts of known concentration from the External RNA Control Consortium (ERCC) were spiked into the samples and the expression values of this RNA was calculated. Accuracy was comparable between the scRNA-seq methods, with Drop-seq having a slightly higher accuracy measure over other techniques.
c. Cost efficiency
The authors calculated the minimal cost for creating sequencing libraries for the number of cells needed for a power of 80% at three sequencing depths, 1, 0.5, and 0.25 million reads. Drop-seq is most cost effective for sequencing at a depth of e.g. 250,000 reads.
The cDNA molecules generated using Smart-seq and Smart-seq2 are not barcoded and instead emphasize full length sequencing coverage of transcripts. Cel-seq, MARS-seq, SCRB-seq, and Drop-seq all produce barcoded cDNA transcripts and add a unique molecular identifier (UMI) sequence. Barcoding and UMI sequences are a particular advantage of these single cell methods and enables tracking the well or cell of origin for transcripts. The use of UMIs improves mRNA quantification because original mRNA molecules can be differentiated from those derived from the amplification step.
In sum, the nature and goals of the experiment will drive the choice of single cell RNA sequencing protocol. In cases with a low cell count, perhaps enriched by FACS, MARS-seq and Smart-seq2 would be advised. Drop-seq allows analysis of a large number of cells and is the most cost-effective for generating sequencing libraries. Therefore, Drop-seq is recommended when trying to identify a particular, small population of cells. In our case, the goal was to identify a rare population of cells (primordial germ cells PGC/small micromeres) in the sea urchin and sea star.
Here we provide detail on two Drop-seq instruments we have utilized, the 10X Chromium (https://www.10xgenomics.com/instrument/) and the Dolomite Nadia instrument (https://www.dolomite-bio.com/product/nadia-instrument/).
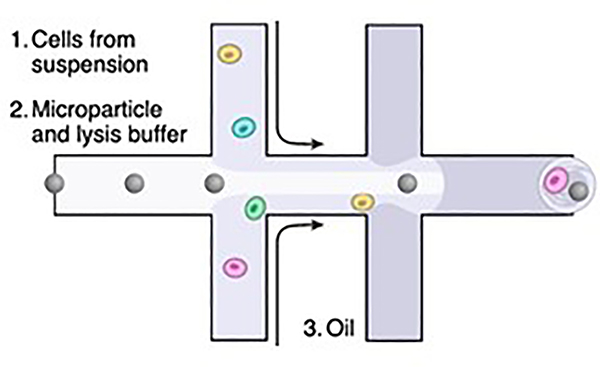
Drop-seq captures single cells by use of a microfluidic chamber. A flow of beads suspended in lysis buffer, a flow of cells, and a flow of oil meet to form thousands of combined emulsion droplets. The beads are coated with oligo-dT primers, which include a PCR priming sequence, a specific barcode unique to each bead, and a unique molecular identifier (UMI). The number of droplets formed greatly outnumbers the number of beads and cells in order to avoid multiplets, cases of more than one bead or cell in a droplet. Once cells are isolated within the droplets, the cells are lysed and the beads capture polyadenylated mRNAs.
d. 10X Chromium
10X Genomics reports approximately 65% cell processing efficiency for the 10X Chromium instrument. The 10X can capture between 500 and 10,000 cells per channel, with a capability of up to 8 channels per run. Cells up to 30μm in diameter have been tested on this device. The microfluidic technology of the 10X Chromium follows that of Macosko et al ((Macosko et al., 2015)), as shown in Figure 1. The resulting gel beads in emulsion are referred to as GEMs.
Figure 1:
Diagram of a generic microfluidic device carrying 1) Cells; 2) microparticles in lysis buffer; 3) oil to separate individual droplets. (from Macosko et al., 2015)
e. Nadia
The Nadia, like the 10X Chromium, is a microfluidic-based system. It has a 10% cell processing efficiency when following recommended concentrations of 300 cells/μL and 600 beads/μL. The Nadia can capture up to 50,000 cells in a single run because it can run a larger volume and a larger number of cells in one run compared to the 10X Chromium. The Nadia can accommodate cell sizes up to 50μm.
Approximately 5,000 cells are generally processed through the 10X Genomics Chromium instrument for embryos. After UMI filtering of the sequencing data, 3,348 (66%) cells are recovered for sequence data analysis. In a comparable Nadia experiment, the recommended concentrations for the cell and bead aliquots are different, but for the same embryo experiments approximately 75,000 cells are run according to manufacturer’s recommendations.
3. Preparation of single cells – tissue dissociation – and CRISPR/Cas9 embryos
a. Obtaining single cells
Dissociating cells from embryos, tissues, or organs requires several important considerations. First, complete dissociation of cells is essential. Doublets, or higher ordered aggregates of cells that go through the drop-seq protocol will be averaged in their transcripts, thereby diminishing the strength of the single-cell procedure. Second, it is essential to have all cells represented proportionally in the dissociation. If the tissue is not completely dissociated, the clumps of cells remaining may represent a distinct population of cells that now will not be present in the analysis. Third, many of the drop-seq protocols can read 5,000 – 10,000 cells per run. If the stage of interest for single cell sequencing is a larva with several thousand cells, and many different cell types, each of them may not be represented at sufficient depth in the final analysis. In such cases it is important to perform multiple runs of the same embryos, and/or to enrich for cells of interest. For example, if one were interested in pigment cells in the ectoderm of a sea urchin larva, one can dissociate the ectoderm off the larvae, spin out the remainder of the larvae, and drop-seq only the separated ectodermal cells. In this case one has decreased the diversity of cell types, and enriched many fold the cells of interest. Where possible, fluorescence activated cell sorting is also of great utility, but it does require specific fluorescence labeling of the cells of interest (Barsi, Tu, Calestani, & Davidson, 2015; Swartz et al., 2014). Protocols for differential dissociation of embryos are documented in this series of manuals (see Chapter 10, Volume 150, D.R. McClay; see also Vol 150 Part A, Chapter 16 (Smith et al) for isolation of adult coelomocytes).
b. Embryos vs larvae vs adult tissues
Many drop-seq instruments and microfluidic chips can handle cells upwards of 40 – 50 microns in diameter. Larger cells, such as eggs or early blastomeres, however, will get clogged in the chip and abort the run. By morula stage and later in many echinoderm species, each cell is less than ~25 microns and is able to flow through a microfluidic device. If the sample of interest is prior to hatching, make sure to treat the embryos at fertilization to inhibit crosslinking of the fertilization envelope so that the embryo can be dissociated. The protocol for this is detailed in Chapter 1, volume 150 (Adams et al), and usually uses 2mM 3-aminotriazole or 10mM p-aminobenzoic acid (pH7.8). Stripping the non-crosslinked envelopes off the embryos usually involves pouring the embryos through a nylon mesh, the size of which is just smaller than the size of the embryos e.g. 64 micron mesh for embryos of S. purpuratus ~80 microns.
To dissociate embryos, larvae and even some adult tissues, we use procedures based on treatments with calcium – free media, followed by gentle dissociation using a loose fitting Dounce homogenizer. For S. purpupratus embryos and early larvae we use the following protocol:
All procedures are on ice, using media that has been stored at 4°C and is kept on ice during the dissociation.
Wash embryos 2x with CaFSW. Pellet embryos using either settling at 1× g, centrifugation in a hand centrifuge for 15 seconds, or 8 seconds in a microfuge (for small numbers e.g. <1000; simply push the manual start button on the microfuge and release it after 8 seconds. The speed will gradually increase from 0 rpm to approximately 8,000 during this time). Aspirate off media and repeat, resuspending embryos each time, and filling the tube each time with new media.
Resuspend embryos in hyalin extraction media (HEM; see below for recipe) for 10 minutes on ice, for embryos prior to gastrulation, 20 minutes on ice, for later embryos (post-gastrulation and larvae), inverting every few minutes.
Pellet embryos and resuspend in 1–2 mls of 0.5M NaCl. The exact volume will depend on the number of embryos and the concentration of cells needed for the drop-seq analysis. This level of this salt does not appear to alter subsequent steps in the procedures.
Place the resuspended embryos in a cold Dounce homogenizer and if possible, perform the douncing under a stereo microscope, watching the effectiveness of dissociation with each stroke. If visualizing under the microscope directly in the Dounce tube is not possible, take a small sample out after a few strokes and visualize embryos and dissociated cells on a microscope slide. Cease douncing when the vast majority (>80%) of embryos have been dissociated.
Pour the dissociated cells through a 40 micron cell strainer to remove any clumps of cells remaining, and evaluate and count the single cells on a heamocytometer. Depending on the drop-seq controller used, cell concentrations in the range of 300/microliter are targeted. If you need to concentrate the cells, spin in a microfuge tube for 12 seconds (as above, from manually starting) and resuspend the cell pellet in 0.5 M NaCl to the desired concentration. In general, pellet the single cells as little as possible in order to minimize breakage of cells in a population that may be more fragile than others, and thereby skewing the dataset.
Although embryos and early larvae from sea urchins dissociate well with calcium-free protocols, some adult tissues do not dissociate to single cells effectively using only this approach. Many mammalian tissues are dissociated using trypsin treatment, and similar approaches work well for echinoderms (e.g. adult ovaries). We have used 0.2% trypsin (use trypsin formulations made for tissue dissociations which are less pure, less expensive, and may have collagenase as well) on ovaries from the sea star Patiria miniata.
Resuspend trypsin to 0.2% in 0.5M NaCl. Dissect ovary and mince it in CaFSW into small pieces using a scalpel or small scissors. Allow the tissue to settle, remove the CaFSW and add the trypsin solution.
Incubate the minced tissue with the trypsin at room temperature for 20 minutes, with gentle mixing.
Settle the tissue, and aspirate off the trypsin solution. Resuspend in 0.5 M NaCl and homogenize with a loose fitting Dounce homogenizer.
We have also used 0.5M urea (really) to dissociate embryos and ovaries. Make 1M urea in distilled water, and dilute 1:1 with CaFSW at 4°C. Add embryos or adult tissues to the urea mix for 10 minutes (4°C) and then replace the media with 0.5M NaCl. Homogenize gently with a Dounce homogenizer as described above.
Note: the reverse transcriptase and cDNA synthesis steps require magnesium ions. Therefore, do not use EDTA in the dissociation steps, only EGTA.
c. Single cell analysis from perturbation experiments
A major value of single cell mRNA – seq is to test for changes in transcript profiles following a perturbation. Some perturbations may be highly impactful on certain lineages, whereas other cell types may be insensitive to the same perturbant. Whole embryo transcriptome analysis, or qPCR analysis from whole embryos would minimize these differences, whereas single cell mRNA-seq would likely reveal these differences and define the cells influenced the most. The perturbation may be from small molecule/drug inhibitors, from environmental changes of the culture condition, from morpholino antisense oligonucleotide (MASO) knock-downs, and/or from CRISPR/Cas9 gene targeting experiments. The conditions for dissociating embryos remain the same as described above. For small molecule inhibitor/drug experiments the numbers of embryos treated are often in vast excess (thousands, generating millions of single cells following dissociation) whereas microinjection experiments often instead yield hundreds of embryos. We found that having ~500 microinjected embryos meant that we would have ample single cells from dissociated embryos, depending on the stage of dissociation. The dissociation of small numbers of embryos is enhanced by observing the entire process under a stereo microscope, both the homogenization and resuspension process.
4. Summary of protocols for obtaining drop-seq emulsions
a. Bead counting
For the Nadia protocol, the first step is to order the barcoded beads from ChemGenes (Catalog number: Macosko-2011–10(V+)). These beads are expensive ($3,524US for 10 umole), and it takes about a month to receive them after placing the order. However, this amount of beads will be enough to process around 50 drop-seq experiments. When the beads are received, they need to be kept at 4°C until use. It is important to use low retention tips (ThermoFisher) during the entire experiment to avoid losing beads, RNA or cDNA. The investigator needs to wash the beads one time with 1 ml of 100 % ethanol plus 1 ml of TE/Tween (10mM Tris pH8, 1mM EDTA, 0.01% Tween) in nuclease free water. This mix is transferred to a 50 ml falcon tube and spun for 1 minute at 1000 g at 4°C. The supernatant is discarded and the beads are washed 2 times with 30 ml of 100% ethanol (using the previous centrifuge settings), and 2 times with 30 ml of TE/Tween. After adding the TE/Tween for the second time, the beads are kept in this 30 ml suspension by either pipetting or inverting the tube, and 20 ul of this mix can be loaded to a Fuchs Rosenthal C-chip haemocytometer to count the beads. It’s important to count at least 4 different regions of the chip to have a more accurate average (the chip contains 16 of these regions). As an example, users here obtained an average of 52 beads per region of the Fuchs Rosenthal. This number is multiplied by 16 to represent the number of beads in the entire Fuchs Rosenthal grid. This grid volume is equivalent to 3.2 ul. To obtain the number of beads per ul, the investigator needs to divide his total number per 3.2. Using the previous example: 52 × 16 = 832 beads in the grid, and 832 beads / 3.2 ul = 260 beads per ul. The stock of beads then contains 30 ml of a solution at a concentration of 260 beads per ul, meaning a total of 7.8 million of beads. This solution has to be aliquoted in Eppendorf tubes of 150 000 beads (number required for each single cell sample). In our example, the investigator will obtain 51 aliquots (150 000 beads / 260 beads per ul = 577 ul per aliquot). These aliquots can be stored at 4°C for several months.
For the 10x Chromium protocol, the beads come with the kit, ready to be used. They only need to be kept at room temperature for 30 minutes before loading the chip.
b. Bead preparation
For the Nadia, on the day of the experiment, the bead aliquot is centrifuged for 1 minute at 1000 g at 4°C. The supernatant is carefully removed (with a p200 followed by a p10 pipette) without touching the pellet, and the beads are then re-suspended in 250 ul of cold Lysis buffer. The Lysis buffer is described in the Nadia protocol (https://www.dolomite-bio.com/support/downloads/). We recommend to make this buffer in advance in a large volume (e.g. 25ml), without DTT, filter it through 0.2 uM filter and store it at 4°C; it will be good to use for several weeks. The DTT needs to be added fresh on the day of the run.
c. Cell Encapsulation
The Nadia instrument is more flexible to use than the 10x Chromium. The Nadia cartridges need to be bought separately, and they are sold as either 1, 2, 4 or 8 identical chips per cartridge. Even though it seems more efficient to directly process 8 samples at once, it requires one to have all the dissociated cell samples on the same day, and more time will be spent in each step to process all of them at once. To improve the quality of the samples, the investigator should use the 1, 2 or 4 chip cartridges. It’s also important to notice that these cartridges are expensive and cannot be reused. If the investigator only needs one chip in a 2-chip cartridge, the second chip is then lost. A set of cartridges for 8 samples, including 2 cartridges of 2 chips and one cartridge of 4 chips costs about 1,500 dollars. For the 10x Chromium, 8 chip cartridges are included in the kit, but unused chips are lost if the investigator only needs to use 4 chips out of the 8 in one run for example.
For the Nadia, all the instructions appear on the screen at the required time. The first step is to add 3 ml of oil (Biorad 1864006) in the oil compartment. The Nadia will then cool down for a few minutes. Following the on-screen instructions, mix and load 250 ul of beads (previously re-suspended in the lysis buffer) and 250 ul of the cells (previously prepared) in their respective compartments (use the p200 and load 2 times 125 ul for both the beads and the cells). The Nadia will form droplets for about 20 minutes at 6 °C, followed by 10 minutes at room temperature to improve cell lysis. The default parameters of the Nadia work well for sea urchin and sea star dissociated cells. However, an additional device from Dolomite, the Nadia Innovate, enables the investigator to control diverse parameters (such as droplet size, droplet frequency, and temperature) and to take live videos of the droplet formation. This device is especially helpful if a sample constantly results in an abnormal emulsion. The user will be able to change the parameters and observe the droplets in the same time. A run with the Nadia takes 28 minutes, with more cells run and an extended lysis step, whereas the 10x Chromium Controller takes 6.5 minutes for fewer cells and no additional lysis incubation.
The resulting emulsion should look dense and white with both instruments. With the Nadia, a green light above the well will indicate to the user which compartment contains the emulsion. The investigator will discard the underlying oil (about 2 ml), and use a low retention tip (as a reminder) to pipette the remaining 1 ml that contains the droplets and transfer it to a low DNA binding tube. It is not a problem if some left over oil is also added to this tube. The beads will be washed in the following steps. Before processing the samples, it is crucial to evaluate the quality of the emulsions under a microscope. 8.5 ul of each emulsion can be loaded into a Neubauer haemocytometer. Droplets and beads will be visible, but the cells have already been lysed. Sometimes, two beads can be found in one droplet but this should be much less abundant than the single bead droplet.
For both instruments, if the emulsion does not look dense or white, there is likely a failure, and the samples need to be run again. If the emulsion looks good but in lower volume than expected, the system probably clogged during the run. The investigator can then decide to re-run the sample, or to process this sample normally especially if the samples are precious.
d. Bead isolation
With the Nadia, the next step is to extract the beads from the droplets. The emulsion is carefully added to the center of a falcon tube containing 30 ml of 6xSSC (Sigma 20x SSC, S6639–1L) diluted in nuclease free water. After addition of perfluorooctanol (Sigma 370533), the tube has to be manually shaken vigorously 3 to 4 times in order to break the droplets. This falcon tube is then centrifuged for 1 minute at 1000 g at 4°C and kept on ice. The isolated beads will be sitting on top of the oil fraction. The supernatant (SSC 6x) can be removed carefully using a 10 ml pipette with a pipette controller. It is better to leave a few ml on top of the beads instead of getting too close to the beads with the pipette. Using the same pipette controller, with a 25 ml pipette, 30 ml of SSC 6x can be strongly released in the same tube to re-suspend the beads. The oil will fraction itself back to the bottom of the tube. This entire bead suspension needs to be quickly pipetted back to a new 50 ml falcon tube using a 10 ml pipette. If this step is not performed fast enough, the beads will start to settle back on top of the oil. It is important to avoid pipetting oil in this step because it will prevent the beads from forming a nice pellet in the next steps. This bead suspension can be centrifuged for 1 minute at 1000 g at 4°C to obtain a pellet of beads. After removing most of the supernatant, the beads are then transferred to a 1.5 ml low DNA bind tube and washed 2 times with SSC 6x (1 minute at 1000 g at 4°C). The beads can be gently re-suspended between these 2 washes, by pipetting them 5 times up and down. With the 10x Chromium, the emulsion is directly used for reverse transcription without isolating the beads.
e. Reverse transcription
For the Nadia, 300 ul of the 5x Reverse Transcription (RT) buffer are added to the pelleted beads to equilibrate them before starting the reaction. They can be re-suspended again by pipetting 5 times up and down before centrifugation. The Reverse Transcription Buffer (made according to the Nadia handbook) can be prepared during the droplet formation in the Nadia and kept on ice until this step is reached. All the components can be mixed and kept on ice, except the reverse transcriptase itself that needs to be added right before the reaction. The reverse transcription will require an incubation of 30 minutes at room temperature followed by 90 minutes at 42°C. It is important in both of these incubations to maintain the beads in suspension (if they settle down, this will prevent the complete reaction). It is recommended to use a disk-style or a rotisserie-style rotator. This protocol requires an unusually high amount of enzyme, and a regular tube of reverse transcriptase (ThermoFisher scientific, EP0751) will not even be enough for one reaction. We advise investigators to buy the large tube right away (EP0753) and to evaluate the left over volume before starting a new run on the Nadia. It is crucial to do the reverse transcription right after the run to avoid RNA degradation. The investigator can then wash the beads according to the Nadia protocol and store the tubes at 4°C overnight.
For the 10X Chromium, the emulsions are incubated at 53°C for 45 minutes, and at 85°C for 5 minutes. The tubes can be stored at 4°C for up to 3 days, or −20°C for a week. In contrast to the Nadia, the bead isolation happens after the reverse transcription in this case. 125 ul of the recovery agent, used to disrupt the droplets is added to each tube. After 60 seconds, and without mixing, a biphasic mixture will appear. The pink fraction contains the recovery agent and the oil, the clear fraction contains the cDNA. After removing most of the pink fraction, without disrupting the clear fraction, 200 ul of Dynabeads Clean up mix are added to the clear fraction. The cDNA is then eluted according to the 10x Chromium protocol (https://www.10xgenomics.com/).
f. Exonuclease treatment
This step is specific to the Nadia instrument. Some beads may have not been in contact with a cell in some droplets, and have not captured any RNA molecule. This exonuclease treatment will remove the primers on these empty beads. The beads are washed with 1 ml of 10 mM Tris pH 8, mixed by pipetting 5 times up and down and centrifuged for 1 minute at 1000 g at 4°C. The bead pellet is re-suspended in 200 ul of exonuclease mix: 22 ul of exonuclease I buffer, 11 ul of exonuclease (New England Biolabs, M0293L) and 187 ul of nuclease free water per sample. (Prepare the mix first in a new tube, the volumes indicated here assume 10% extra volume for pipetting losses. Then pipette 200 ul of this mix to the tube containing the beads). After 45 minutes at 37°C, the beads are washed one time with TE/SDS (10 mM Tris pH 8, 1 mM EDTA pH 8, 0.5% SDS) and one time with TE/TW.
g. Summary of protocols for making sequencing libraries
PCR
For the Nadia instrument, before making the cDNA library, the beads need to be washed 2 times with 1 ml of nuclease free water, and counted using a Fuchs-Rosenthal haemocytometer (as previously described in the bead counting section). The average number of beads per region (for example: 14) will be multiplied by 16 and divided by 3.2 to obtain the number of beads per ul (for example: 14×16/3.2= 70). The beads were previously re-suspended in 1 ml of nuclease free water, giving a total number of beads of 70 × 1,000 ul = 70,000 beads per ul in our example. The Nadia protocol recommends to adjust the bead concentration to 200 beads per ul and to use 10 ul of this mix (2,000 beads) to make the cDNA library for each sample. In this protocol, the left over (68,000) beads will be lost and rare cell types might also be lost if they were not represented in the chosen 2,000 bead aliquot. We suggest to amplify the entire amount of beads to obtain a more accurate representation of the sample. In this case, the beads can be diluted to 500 beads per ul instead of 200, resulting in 5,000 beads per PCR reaction. Instead of doing a single PCR per sample, we suggest to aliquot all the beads into PCR tubes (for example, 70,000 / 5,000 = 14 PCRs for one sample). These PCRs will be pooled together in one single tube at the end of the reaction, following their purification with the AMPure XP beads (Beckman Coulter, A63880) as explained in the Nadia protocol.
For the 10X Chromium, the entire volume of purified cDNA (35 ul) is amplified in one single tube after addition of 65 ul of the Amplification reaction mix. It is important to use a Thermo cycler that can accommodate a reaction volume of 100 ul. This PCR is then washed with the SPRIselect reagent. For both instruments, the quality of the amplified DNA need to be tested with the Fragment bioanalyzer (size and concentration of the DNA).
Tagmentation
The fragments resulting from the PCR are expected to be larger than 1kb, since they often represent the full length cDNA. However, these fragments cannot be directly sent for Illumina sequencing. They need to be cleaved in smaller fragments and tagged for sequencing purpose in order to be sequenced on an Illumina flow cell. Fragments larger than ~700 bps cause cluster overlap, and decreased signal, reducing the sequence efficiency.
The Nadia protocol suggests using the Nextera XT kit (Illumina, FC-131–1024) for the tagmentation. Only 600 pg of full length cDNA will be tagmented for each sample. The Nextera XT kit relies on the transposase Tn5 that will cleave the DNA and tag it with adaptors. However, in our hands, this Nextera XT kit does not lead to a sufficient amount of DNA for sequencing purposes and it required trouble shooting for each sample in order to obtain the correct fragmentation size. A new kit type is now available, the Nextera Flex (20018704); it delivers consistent insert sizes regardless of the DNA input amount or size. This new bead-based technology minimizes bias and opportunities for error, resulting in highly reproducible sequencing data. We recently tested this new Nextera Flex kit, and obtained perfect fragmentation for all our samples. Moreover, this kit enables investigators to fragment larger amounts of full length cDNA (1 ng to more than 500 ng) and to obtain a reproducible size of fragments (~500 bps). We strongly recommend the use of the Nextera Flex kit since it also leads to tagmented libraries that are about 10 times more concentrated than with the Nextera XT kit.
The 10x Chromium samples are fragmented with a specific fragmentation buffer included in the kit. In contrast to the Nadia, here almost the entire sample is tagmented (35 ul out of the 40 ul eluted in the previous step). This fragmentation step is followed by a purification with the SPRIselect reagent. The adaptors required for the Illumina sequencing are then ligated to the fragments. These ligated fragments are purified once again with the SPRI reagent.
Sample Indexing
Before sending the samples for sequencing, they need to be indexed. These indexes are especially important if the investigator decides to run several libraries in the same sequencing lane. The samples that will be run together need to have a different index, and the samples run separately can have the same index. The indexes will be used to identify the origin of the transcripts. For the Nadia, the samples can be indexed using the Nextera XT index kit (Illumina, FC-131–1001) if the libraries were made using the Nextera XT kit or using the Nextera DNA CD index kit if the Nextera Flex kit was used (Illumina, 20018707). It is not recommended to use XT indexes with Flex libraries as Nextera DNA CD kit has indexes of higher purity that have been carefully optimized for improved performance. The indexes are added to the tagmented libraries by PCR. With the Nextera XT index kit (presented in the Nadia handbook), only one variable index (N7XX) is added to each library. The second one is constant in each library (using the New P5 smart PCR hybrid oligo). Each PCR is purified with the previously used AMPure XP beads and analyzed on the Fragment bioanalyzer. With the Nextera DNA CD kit, two variable indexes are added by PCR; the indexed DNA is then purified using the purification beads provided in the kit.
For the 10x Chromium, the reagents are provided in the kit. Only one variable index is also added in each library (I7 index). Each PCR is purified with the previously used SPRIselect reagent and analyzed on the Fragment bioanalyzer. In each case, the fragments should now be around 500 bp.
The 10x Chromium offers an additional step to better evaluate the library concentration. In addition to the concentration obtained with the Fragment bioanalyzer, the investigator can also evaluate the library concentrations using qPCR (https://www.10xgenomics.com/).
Sequencing
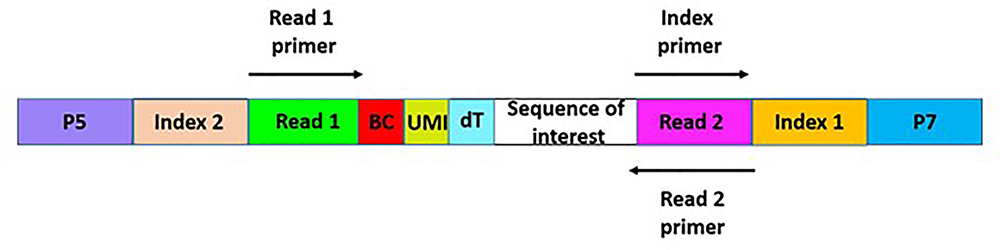
Regardless of the construction methods described above, the libraries contain the sequence of interest surrounded by adapter constructs on each side (Figure 3). Illumina generally asks libraries to be submitted at 15 nM in 20 ul for the HiSeq2500 instrument. An in-depth view of the transcriptome will require about 200 million reads per sample.
Figure 3:
Map of the cDNA product from drop-seq. Adapters have the flow cell binding sites, P5 and P7, to allow the library to attach to the Illumina flow cell surface. A primer complementary to Read 1 will allow the sequencing of the bead barcode (BC), as well as the UMI (Unique Molecular Identifier). When Read 1 is finished, everything from Read 1 is removed and an index primer is added. A primer complementary to Read 2 is then used to sequence the cDNA fragment. Paired end sequencing is essential to obtain these sequences: 26bp read1, 8bp sample index, 98bp read2.
5. Analysis of scRNA-seq results: general considerations
Several recent reviews outline procedures and challenges with analyzing scRNA-seq data and provide recommendations regarding specific tools for each stage of analysis ((Stegle, Teichmann, & Marioni, 2015) (Poirion, Zhu, Ching, & Garmire, 2016) (Haque, Engel, Teichmann, & Lonnberg, 2017) (Dal Molin & Di Camillo, 2018)). Here we discuss general issues regarding analysis of scRNA-seq results as well as points particular to studying echinoderm development. The following section covers a few specific workflows.
Although scRNA-seq and bulk RNA-seq analyses are similar in many regards, there are some critical differences ((Stegle et al., 2015) (Dal Molin & Di Camillo, 2018)). A point to keep in mind from the outset is that scRNA-seq provides a powerful way to identify distinct regulatory states within complex cell populations, whereas bulk RNA-seq is generally better for measuring quantitative differences in gene expression. Thus, single cell and bulk analysis pipelines are often designed to provide different biological insights ((Stegle et al., 2015) (Griffiths, Scialdone, & Marioni, 2018)). A common mistake is expecting scRNA-seq data to address questions that may be better tackled using bulk RNA-seq, such as measuring changes in transcripts across development. Both approaches are useful, but neither is the best in all situations.
An operational challenge in analyzing scRNA-seq data is that procedures are not yet standardized, due to the rapid evolution of the technologies that produce the data. Improved analytical practices and implementations are appearing rapidly, so it is important not to “clone” analysis pipelines directly and uncritically from previously published studies but rather to use them as a starting point and then consult the literature for subsequent improvements. Another general point is that methods of analysis may need to be modified depending upon the specific scRNA-seq technology. In particular, droplet- and plate-based approaches generate very different read depth per cell from (usually) very different numbers of cells, which needs to be taken into account during analysis.
Two intrinsic limitations with scRNA-seq data should be kept in mind. One is the relatively low number of reads that can be obtained from a single cell, which is a direct consequence of the number of template molecules and the generally low efficiency of loading onto reverse transcriptase. Given the complexity of most transcriptomes and the strongly skewed frequency distribution of transcripts per gene, many genes with low to moderate expression will not be represented by any reads for any given cell. This phenomenon, known as read drop-out, limits the sensitivity of scRNA-seq and introduces challenges for statistical comparisons among samples, such as detecting and quantifying expression differences ((Hicks, Townes, Teng, & Irizarry, 2018)).
The other intrinsic limitation to be aware of is that amplification of the initial cDNA templates distorts relative read abundances, especially for moderate to low abundance transcripts which, in most cases, will comprise a minority of all reads but the vast majority of distinct transcripts. As a result, the apparent rank order of transcript abundances in a given cell may not accurately reflect the situation in the live cell. Using single molecule-level unique molecular identifiers (UMIs) ((Islam et al., 2014)) provides a useful way to improve quantitation. Note that it is not possible to incorporate UMIs with some single-cell library preparation methods. The other way to improve accuracy is to include information from more cells per sample. Producing large suspensions of synchronous, healthy, single cells is relatively straightforward when working with echinoderm embryos and lavae ((McClay, 2004)). Preparing a viable single cell suspension is essential for obtaining biologically meaningful results from scRNA-seq data, and is much more challenging with some other animal model organisms, for instance due to the presence of a cuticle.
With scRNA-seq, it is important to optimize informative read alignment, because the number of reads is limiting. Alignment of ~40–50% of reads is common, even when working with a well annotated reference genome ((Ziegenhain et al., 2017)), which is a lower proportion than generally achieved with bulk RNA-seq. Working with echinoderm samples introduces a further challenge to alignment, because the high level of genetic variation present in most populations can result in reads being discarded because they contain SNPs that are not represented in the reference genome. Most mapping programs allow the user to alter the number of allowable mismatches. Be aware that the default setting may be based on organisms with less overall genetic variation than echinoderms. Mapping efficiency and the number reads mapping to multiple locations will both rise as the number of allowable mismatches increases, making this an important parameter to examine critically. Because the former provides better coverage while the latter reduces it, use a setting that maximizes mapping efficiency to a single location. An empirical solution to this problem is to split samples and carry out bulk RNA-seq in parallel with scRNA-seq. The bulk reads can then be used to construct a cross-specific reference transcriptome that largely factors out genetic variation when mapping scRNA-seq reads.
6. Analysis procedures: Overview of procedure pipelines for analysis of results
a. Partek Flow
Partek, offers a robust sequencing analysis software, comparable to that of 10X Genomics (below), called Partek Flow [http://www.partek.com/partek-flow/]. Besides the Linux based system, Partek Flow can be accessed through a browser based interface after purchasing a license hosted on their server. Hosting your license on a Partek server grants you the ability to house your data on their servers and the data can easily be downloaded from the server. The drop-seq pipeline accessed through Partek Flow follows the same steps as depicted for the 10X sequence analysis pipelines. A noteworthy difference between the two software applications is that while the 10X Cell Ranger is optimized to work with 10X data, Partek Flow can analyze both 10X, Nadia, and other datasets in addition to hosting modules for RNA-seq, ChIP-seq, microarray analysis, and others. Partek emphasizes Partek Flow’s ease of use, particularly that pipelines can be run with little coding knowledge. For users seeking more customization, Partek Flow allows you to adjust the scripts developed for each analysis step.
b. Expression analysis using Cell Ranger (10X Genomics R package)
10X Genomics offers several software pipelines for analysis of sequencing data. The Cell Ranger Single Cell Software Suite 2.1.0, run on Linux, demultiplexes the Illumina base call files (BCL), the file containing the nucleotide base calls for each sequencing cycle, into FastQ files, which contain both the base calls and a quality measure for all reads. Cell Ranger also performs UMI counting. In our case, Cell Ranger was also used to generate the S.purpuratus reference genome. Secondary analysis of gene expression data can be accomplished using Cell Ranger or using R or Python. Clustering analysis for these samples was accomplished using Seurat, an R package for scRNA-seq analysis developed by the Satija Lab [https://satijalab.org/].
The first step in the Cell Ranger analysis pipeline is cellranger mkfastq, which produces the FASTQ files. Cellranger count performs genome alignment and produces UMI counts in the form of a matrix, this is done individually for each sample. Cellranger aggr combines the matrices from individual runs of cellranger count to normalize the data based on sequencing depth. This is an essential step in creating a gene-barcode matrix for an entire experiment.
The gene-barcode matrix file produced can be uploaded to R for downstream analysis. The first column in the matrix is the list of genes that were sequenced and mapped. In the row for one gene, the information that follows is the barcode that was linked to that gene transcript.
LOC585686 TTTGGTTAGAACTCGG TTTGGTTAGCTCAACT TTTGGTTAGTACGCGA
LOC764781 TTTGGTTAGTGACATA TTTGGTTGTACCAGTT TTTGGTTGTATAGTAG
LOC579747 TTTGGTTTCACCCTCA TTTGGTTTCCAAATGC TTTGGTTTCCACGACG
Cellranger reanalyze is a pipeline that allows you to change the default parameters for visualizing the gene expression data of your experiment. See [https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome] for details and flow of the procedures for 10x Chromium datasets.
Loupe Cell Browser, a 10X Genomics application, is used to visualize the clustering and gene expression data output from running the Cell Ranger pipelines. Cell Ranger pipeline output files contain results for t-SNE projection, principal component analysis (PCA) and k means clustering. K means clustering assigns cells to k number of groups based on their characteristics. This form of clustering is helpful when the identity of the cell types, in terms of gene expression, is unknown. The 10X Genomics website offers a tutorial (PBMC data), which walks the user through seeing the pipeline results in R: http://cf.10xgenomics.com/supp/cell-exp/cellrangerrkit-PBMC-vignette-knitr-2.0.0.pdf.
c. Expression Analysis Using Seurat (platform agnositic R package)
The Satija Lab offers different tutorials for the Seurat R package based on the nature of the experiment. The tutorial titled, “Integrating stimulated versus control PBMC datasets to learn cell type specific responses” (https://satijalab.org/seurat/immune_alignment.html) matches the sea urchin experiment best when the goal is to analyze changes in particular cell types between the control and treated samples or compare gene expression changes in particular cell types over time. We used this code for our analysis with a few adjustments to cater to the echinoderm system.
The first step in the analysis using Seurat is quality control, or QC. The QC step includes filtering cells and normalizing the data. In this line of code, the cells within the 10X data stored in the variable “C1” are filtered based on numbers of genes.
As part of the QC, we filtered cells by setting a low threshold of 400 genes and a high threshold of 2500 genes. These thresholds are set to exclude droplets where a complete cell was not captured and multiplets, where more than one cell lysed in a droplet would present as having many more genes.
Following the given code, we are able to find variable genes in the dataset and employ the RunTSNE command. Creating a principal component analysis plot is common for analysis of single cell sequencing data. The use of canonical correlation analysis (CCA) is similar to the of principal component analysis, both of these analyses look for sources of variation between your control and experimental datasets. The Seurat tutorial includes a CCA step but others may make use of PCA plots.
d. Principal Component Analysis (PCA)
Principal Component Analysis (PCA) plots are based on variation in the sequencing data with the first principal component representing the greatest variation among samples, the second the next most, and so forth; the loading for each principal component quantifies how much of the variation it explains. You can imagine a case where this would be one experimental condition against the control so the PCA is originally plotted multidimensionally.
There are numerous resources for understanding PCA plots and an interactive example may be helpful in seeing how variability in the dataset affects a 3D plot. Analyzing a multidimensional plot poses its own challenges, however, and reducing the data to two dimensions can help with visualization.
An interactive PCA tutorial is available online: http://setosa.io/ev/principal-component-analysis/
After choosing the appropriate correlation values for analysis, the tutorial walks the user through generating t-distributed stochastic neighbor embedding plots.
e. T-Distributed Stochastic Neighbor Embedding
T-Distributed Stochastic Neighbor Embedding (t-SNE) plots provide an alternative way to visualize clusters of cells in scRNA-seq data. Given the sparse nature of count tables with scRNA-seq data, where many cells contain zero counts, t-SNE is a more appropriate clustering algorithm than PCA. Each point on a t-SNE plot is representative of one cell, defined by two t-SNE components. Proximity on the two dimensional plot corresponds to similarity in the gene expression profiles of the cells. Clusters therefore correspond to a group of cells with similar expression and likely function. The identity of the clusters must be drawn from the unique gene profiles of the clusters. The expression of marker genes can also be shown on a t-SNE plot—this can be shown as expression greater than zero or as a range.
The FindClusters command, which is employed after the RunTSNE command, has a “resolution” parameter. Setting resolution to a value greater than 1 will give more clusters in the t-SNE plot, and a value below 1 will give fewer. While this can be modulated, the goal is to visualize distinct, unique cell clusters.
Once the clusters are visualized on a t-SNE plot, Seurat can extract what combinations of genes are characteristic of each cluster of cells, based on over- or under-representation of reads among all cells in the cluster. The job of identifying cell types either based on the presence of particular genes, sets of genes, or overrepresentation analysis begins here.
f. Gene Ontology
Gene Ontology (GO) term enrichment is used to interpret sets of genes based on annotations for the reference gene set. Once you have compiled your list of conserved and differentially expressed genes for each cluster, you can use this list for overrepresentation analysis. The S.purpuratus reference is present on the GO website. There is also information on how to interpret results on the GO website: http://geneontology.org/page/go-enrichment-analysis.
g. Cell lineage and pseudotime
Seurat also provides the ability to reconstruct cell lineages from scRNA-seq data. For embryonic development, samples need to be collected in a time series to represent changes in gene regulation, whereas for stem cell populations that are continuously producing differentiated cells, a single time point is sufficient. In either case, Seurat can reconstruct the branching process of regulatory states and highlight the specific transcripts that differentiate them. It can also assign pseudotime values that places each cell into an inferred location within the overall series process.
7. Alternative single cell technologies
The technology in single cell RNA sequencing is changing rapidly. On the horizon (late-2018) is sequencing of RNA from nuclei (not cytoplasmic) using drop-seq technologies, so called Single nuclei RNA-sequencing (DRONC-seq). This is particularly useful for tissues that are extremely hard to dissociate (e.g. mammalian brain tissue) and in which the cells are lysed or damaged in the dissociation process. This is less essential for cells that instead dissociate readily as in echinoderms, but the technology could help investigators distinguish between mature mRNA and newly transcribed RNAs for example, or in characterizing tissues from adult e.g. tube feet that might be difficult to dissociate. The 10x company also announced recently an effort for single cell ATAC-seq, to identify open chromatin sites in genomic DNA on a per cell basis. Such an experiment could reveal the heterogeneity in cellular function or fate, even before the phenotype was apparent. Hopefully, the echinoderm community is quickly able to make use of these approaches to study important biological functions, and to take advantage of the many wonderful characteristics that these embryos and animals have to offer.
B. Nascent RNA capture
Many procedures for studying embryonic development rely on testing steady state levels of mRNAs. That is, we normally measure the resulting balance of mRNA synthesis with mRNA degradation. These assays usually consist of in situ hybridization, qPCR, RNA seq, or single cell transcriptome analysis for example. Each mRNA, however, has its own dynamics. Some mRNAs have a rapid turnover, so that the investigator is measuring genes recently transcribed, whereas other mRNAs can be prolonged in their half-life, misleading the investigator into thinking the gene was transcriptionally active at the time of the measurement. The protocol here documents how investigators test for newly transcribed mRNAs and how to distinguish them from aged, long-lived transcripts.
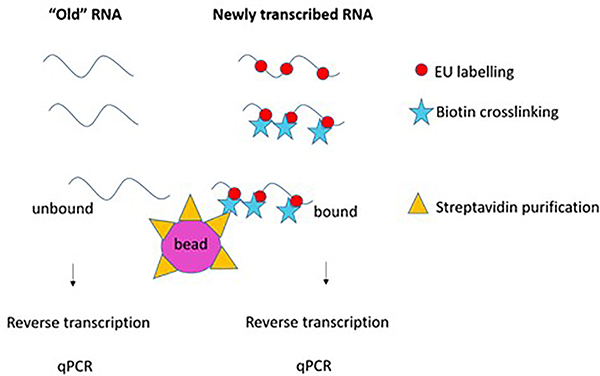
The Click-iT Nascent RNA Capture Kit (ThermoFisher scientific, C10365) employs a modified nucleotide that is incorporated into growing, nascent mRNAs. The nucleotide analog is tagged so that the investigator can then measure how much transcript was made during the incubation window of time. RNA expression also changes with specific perturbations (protein inhibitors, morpholinos, CRISPR, etc.), this nascent RNA procedure is used to determine the effects of these perturbations on specific, or general transcriptional activity. Briefly, the newly transcribed RNAs are labelled with EU, crosslinked to Biotin using the Click-It technology, purified with Streptavidin beads, reverse transcribed and tested by qPCR, or RNA-seq (Figure 4).
Figure 4:
Flow of protocol for identifying nascent mRNA expression in cells using EU labeling.
Protocol
The stock of EU is re-suspended in DMSO at a final concentration of 200 mM according to the manufacturer’s instructions.
We suggest performing these incubations in small volumes e.g. 12 well plates (final volume of 2 ml per well) to reduce the volume of EU needed as this reagent is costly. We recommend to use approximately 250 embryos per well.
Add EU to a final concentration of 1mM EU for the desired time: a few hours to several days, depending on the experimental goal ((Fresques & Wessel, 2018)). The time will depend on the abundance of the transcript of interest and the developmental time point(s) that are being analyzed. It is important to note that the shorter the incubation, the better the temporal resolution, but the lower the signal of labeled mRNAs. Embryos do have a large amount of free nucleotides within the cytoplasm ((Davidson, 1976)) and use of the analog is concentration and time dependent.
At the end of the labeling period, the embryos are transferred into Eppendorf tubes and centrifuged for 1 minute at 3000 rpm. Keep tubes and wash solutions on ice to reduce further transcriptional activity and swimming of the embryos back into the supernatant. After the centrifugation, the supernatant is removed and the embryos are re-suspended in 75 μl of RLT buffer provided in the Rneasy micro Kit (Qiagen, 74004). The samples can be stored at −80°C or the RNA can be extracted right away following this Rneasy micro kit protocol. The purified RNA can be eluted in 20 ul of nuclease free water, Nanodropped and stored at −20°C. Unincorporated nucleotides are washed out during the RNA isolation procedure so free nucleotides are not a complication in the labeling procedure.
If there is an interest in measuring the half-life of various transcripts that were transcribed during the labeling period, do the following. After the pulse period of labeling, wash the embryos at least 3 times and add 10mM buffered UTP for the duration of the incubation to compete effectively with residual nucleotide analog.
-
Crosslink the Biotin Azide reagent in the kit to the nascent EU labelled RNA using the click-iT reaction buffers provided. Prepare the Click-iT reaction cocktail according to the protocol. The concentration of Biotin Azide depends on the concentration of the purified RNA as explained in the manufacturer’s protocol. As an example, here is the protocol that we used for most of the samples:
25 ul of Click-iT EU buffer,
4 ul of CuSO4,
2.5 ul of Biotin azide (0.5 mM final),
15.75 ul of EU labelled RNA (at 100 ng/ul corresponding to a total amount of 1.575 ug),
1.25 ul of Click-iT reaction buffer additive 1,
1.5 ul of Click-iT reaction buffer additive 2.
The mix will turn dark brown after addition of the buffer additive 2. The reaction is performed for 30 minutes at room temperature under gentle vortexing. We recommend using a Thermo mixer with the following settings: 350 rpm at 22°C. The RNA (Biotin EU labelled and unlabeled RNA) can then be precipitated and re-suspended in 50 ul of nuclease free water, following the protocol given. Using sea star larvae, we obtained about 30 ng/ul.
-
The Biotin EU labelled RNA then is purified with 15 ul of Dynabeads MyOne Streptavidin T1 magnetic beads (ThermoFisher Scientific, 65601) for each sample. If the investigator has several samples, we recommend washing the total amount of beads (150 ul for 10 samples) all together before aliquoting them into separate reactions. For each sample, the RNA binding mix is prepared as follows:
49 ul of Click-iT RNA binding buffer 2x,
2 ul of RNAseOut Recombinant ribonuclease inhibitor,
49 ul of purified RNA (previously re-suspended in 50 ul but 1 ul was used for nanodrop).
This RNA binding mix is heated at 70°C for 5 minutes before adding 15 ul of bead suspension in each reaction. The binding reaction is performed in the Thermo mixer using the previous parameters (30 minutes at room temperature, ~ 22°C).
The beads are immobilized using the tube magnet. We recommend keeping the unbound fraction (approximately 100 ul) before starting the wash, it represents the RNAs that were already present in the embryos but that were not being transcribed during the time of the EU incubation.
The beads are washed according to the protocol, and re-suspended in 15 ul of the Click-iT reaction wash buffer 2. We suggest to precipitate the 100 ul of unbound fraction (using the previous protocol: glycogen, ammonium acetate and ethanol) and re-suspend it in the same volume as the labelled RNA: 15 ul of nuclease free water.
Both fractions need to be reverse transcribed right away to avoid RNA degradation. We routinely use the Maxima first strand cDNA synthesis kit (ThermoFisher Scientific, K1641) for reverse transcription. Analysis of the cDNA now depends on the experimental goals, and may include sequencing, or specific transcripts can then be analyzed in each fraction, using qPCR. We use Maxima SYBR Green/ROX 2X qPCR Master Mix (ThermoFisher Scientific, FERK0222) for qPCR applications.
C. Chromatin Immunoprecipitation
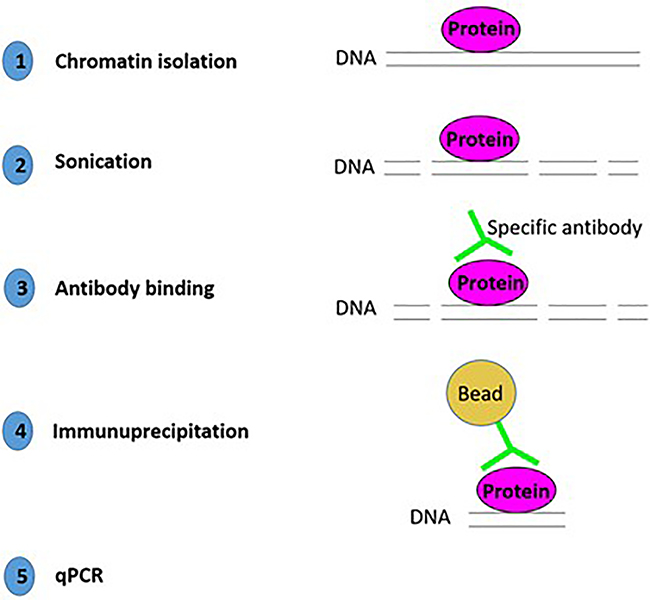
Chromatin Immunoprecipitation (ChIP) is a powerful method to test specific protein - DNA interaction (Figure 5). Briefly, the chromatin is isolated and sonicated to shear the DNA in fragments of approximately 200 bp. An antibody against your favorite DNA binding protein is used to immunoprecipitate the DNA-protein complex. After purification of this complex with magnetic beads, the bound DNA is analysed by qPCR for promoter specific studies or by ChIP-seq to identify new targets. Here, we focus on the ChIP protocol followed by specific qPCR analysis.
Figure 5:
Flow of steps for chromatin immunoprecipitation (ChIP).
Cell dissociation and fixation (based on a protocol used in Drosophila ((Larschan et al., 2007)))
-
1
The embryos are cultured in 50 ml falcon tubes (0.25% v/v, meaning 125 ul of pelleted embryos in 50 ml) until the desired developmental time point.
-
2
After centrifugation, wash the pelleted embryos 3 times with 25 ml of calcium free sea water (CaFSW, see below).
-
3
Re-suspend the embryos in 10 ml of dissociation buffer (1M Glycine, 25mM EDTA, pH 8.0) and keep on ice for 10 minutes (Juliano et al, 2014), then disaggregate them by a loose fitting homogenizer.
-
4
Wash the dissociated cells 3 times with 5 ml of calcium free seawater by centrifugation (1500g for 5 minutes).
-
5
After resuspension, fix these 5 ml of dissociated cells by adding 5 ml of formaldehyde 2% (stock made in calcium free seawater), to obtain a final concentration of 1%.
-
6
Rotate the sample at room temperature for 10 minutes.
-
7
Add glycine to a final concentration of 125 mM, and rotate at room temperature for 5 minutes.
-
8
Transfer the cells to a 15 ml Falcon tube and keep it on ice.
-
9
Spin at 1500g for 5 minutes at 4°C.
-
10
Discard the supernatant, and re-suspend the pellet in 5 ml of PBS− 0.5mM EDTA pH 7.2 + 5 ul of PMSF 0.2 M
-
11
Spin at 1500g for 5 minutes at 4°C.
-
12
Discard the supernatant, and re-suspend the pellet in 1 ml of ChIP wash buffer A.
-
13
Transfer the sample to a 1.5 ml Eppendorf tube.
-
14
Rotate the tube for 10 minutes at 4°C.
-
15
Spin at 1500g for 5 minutes at 4°C.
-
16
Discard the supernatant, and re-suspend the pellet in 1 ml of ChIP wash buffer B
-
17
Rotate the tube for 5 minutes at 4°C.
Cell counting
-
18
Dilute 10 ul of the sample with 90 ul of ChIP buffer B. Continue to rotate at 4°C while counting.
-
19
Count the cells using a haemocytometer.
-
20
Spin the rest of the sample (tube that was rotating), at 1500g for 5 minutes at 4°C.
-
21
Remove the supernatant entirely.
-
22
Freeze the pellet in liquid nitrogen, and store it at − 80°C.
Cell lysis and sonication
-
23
Re-suspend the pellet in 300 ul of ChIP Lysis buffer.
-
24
Lyse the cells by rotating at 4°C for 10 minutes.
-
25
Transfer the sample to a Protein low Bind tube.
-
26
Sonicate with the Bioruptor: 6 rounds of 5 minutes (each round is programmed to alternate between 30 seconds on and 30 seconds off). Add ice in the Bioruptor after each round.
-
27
Spin at 13,000 rpm for 10 minutes at 4°C.
-
28
Transfer supernatant to a 15 ml Falcon tube, and add 9 volumes of ChIP dilution buffer (for example, add 2.7 ml of buffer in 300 ul of sample)
-
29
Remove 400 ul for DNA input, transfer it to a new Protein Low Bind tube.
-
30
The rest of the sample (2.6 ml) is aliquoted into 3 Protein Low Bind tubes (860 ul per tube) and frozen in liquid nitrogen. Store the samples at − 80°C.
Input DNA Clean-up
-
31
To the 400 ul of input DNA, add
21.5 ul of 20% SDS
15 ul of 5M NaCl,
1 ul of RNase A (10/mg/ml stock)
-
32
Reverse crosslink the DNA at 65°C (hot plate) overnight
-
33
On the next day, add 1 ul of RNAse A and incubate at 37°C for 30 minutes.
-
34
Add 20 ul of 1M Tris-HCL pH 6.8
10 ul of 0.5 M EDTA pH 8
3 ul of 20 mg/ml proteinase K
-
35
Incubate at 42°C for 90 minutes
-
36
Perform phenol chloroform isoamyl alcohol extraction
Add 500 ul of phenol chloroform isoamyl alcohol to the sample and shake gently (no vortex)
Spin at 14,000 rpm for 5 minutes at room temperature.
Save the supernatant in a new tube.
-
37
Repeat the extraction with chloroform only
-
38
Precipitate the DNA (about 500 ul of supernatant) with ethanol, add:
10 ul of glycogen
1/10 volume of 3M NaOAc (about 50 ul)
2 volumes of 100 % ethanol (about 1 ml)
-
39
Incubate at − 80°C overnight.
-
40
Spin at 14,000 rpm for 10 minutes at 4°C.
-
41
Discard the supernatant and wash the pellet with 1 ml of fresh cold 70% ethanol
-
42
Spin at 14,000 rpm for 10 minutes at 4°C.
-
43
Discard the supernatant and spin down again for 1 minute to remove the residual ethanol.
-
44
Open lid and leave the tube at room temperature for 1 minute to dry the pellet.
-
45
Re-suspend the pellet in 100 ul of ddH2O
-
46
Nanodrop the sample
-
47
Run 500 ng of the sample on a 2% agarose gel. The DNA should form a smear around 200 bp.
-
48
Store this input DNA sample at − 80°C, it will be used to normalize the qPCR data.
Immunoprecipitation
-
49
Thaw the tubes of diluted chromatin prep on ice
Add your specific antibody at appropriate dilution (usually 1/1000 to 1/500) in one tube. Add control IgG to a second tube at the same concentration as your specific antibody. If you use a rabbit affinity purified antibody, use rabbit affinity purified IgG as a control. Your third aliquot can be used to test a second specific antibody in the same experiment.
-
50
Rotate overnight at 4°C.
-
51
The next day, add 50 ul of magnetic protein A dynabeads to each IP sample (optional: pre wash the beads with 500 ul of ChIP dilution buffer before adding them to the samples)
-
52
Rotate for 2 hours at 4°C.
-
53
Use the magnet to immobilize the beads and discard the supernatant between each of the following washes.
-
54
Wash 2 times with 1 ml of cold RIPA 150. Rotate at 4°C for 5 minutes between each wash.
-
55
Wash 1 time with 1 ml of cold RIPA 300. Rotate at 4°C for 5 minutes.
-
56
Wash 1 time with 1 ml of cold LiCl/TE buffer. Rotate at 4°C for 5 minutes.
-
57
Wash 2 times with 1 ml of cold TE buffer. Rotate at 4°C for 5 minutes between each wash.
-
58
To elute, add 250 ul of Sodium bicarbonate buffer to the beads. Rotate at room temperature for 15 minutes and transfer the supernatant to a new Protein low bind tube.
-
59
Repeat the elution in the same volume, and add the supernatant to the same tube.
-
60
Add 20 ul of 5M NaCl
-
61
Incubate the samples at 65°C overnight.
IP DNA Clean-up
-
62
On the next day, add 1 ul of RNAse A and incubate at 37°C for 30 minutes.
-
63
Add 20 ul of 1M Tris-HCL pH 6.8
10 ul of 0.5 M EDTA pH 8
3 ul of 20 mg/ml proteinase K
-
64
Incubate at 42°C for 90 minutes
-
65
Perform phenol chloroform isoamyl alcohol extraction:
Add 500 ul of phenol chloroform isoamyl alcohol to the sample and shake gently (no vortex)
Spin at 14,000 rpm for 5 minutes at room temperature.
Save the supernatant in a new tube.
-
66
Repeat the extraction with chloroform only
-
67
Precipitate the DNA (about 500 ul of supernatant) with ethanol, add:
10 ul of glycogen
1/10 volume of 3M NaOAc (about 50 ul)
2 volumes of 100 % ethanol (about 1 ml)
-
68
Incubate at − 80°C overnight.
-
69
Spin at 14,000 rpm for 10 minutes at 4°C.
-
70
Discard the supernatant and wash the pellet with 1 ml of fresh cold 70% ethanol
-
71
Spin at 14,000 rpm for 10 minutes at 4°C.
-
72
Discard the supernatant and spin down again for 1 minute to remove the residual ethanol.
-
73
Open lid and leave the tube at room temperature for 1 minute to dry the pellet.
-
74
Re-suspend the pellet in 100 ul of ddH2O
-
75
Nanodrop the samples
qPCR
-
76
Dilute the input DNA to 2 ng/ul and use 1 ul per qPCR control reaction
-
77
Use 1 ul of immune-precipitated DNA per qPCR ChIP reaction
-
78
All qPCR reactions are run in triplicate using the Maxima SYBR green reagent.
-
79
Data is analyzed using the percent input method.
Buffers
ChIP Buffers
The following buffers can be made in advance in 50 ml falcon tubes and filtered through sterile 50 ml disposable vacuum filtration system 0.22 (Millipore, SCGP00525). They can be kept at 4°C for several weeks. The 1x protease inhibitors, and the PMSF need to be added fresh on the day of the experiment.
0.2 M PMSF
0.174g PMSF
5 ml Isopropanol
ChIP wash buffer A
10 mM Hepes pH 7.6
10 mM EDTA pH 8
0.5 mM EGTA pH 8
0.25 % Triton X-100
Filter sterilize
1x protease inhibitor
0.2 mM PMSF
ChIP wash buffer B
10 mM Hepes pH 7.6
100 mM NaCl
1 mM EDTA pH 8
0.5 mM EGTA pH 8
0.01% Triton X-100
Filter sterilize
1x protease inhibitor
0.2 mM PMSF
ChIP lysis buffer
0.1% SDS
10 mM EDTA pH 8
50 mM Tris-HCl pH 8
Filter sterilize
1x protease inhibitor
0.2 mM PMSF
ChIP dilution buffer
0.01% SDS
1.2 mM EDTA pH 8
16.7 mM Tris-HCl pH 8
1.1% Triton X-100
167 mM NaCl
Filter sterilize
1x protease inhibitor
0.2 mM PMSF
ChIP TE wash buffer
10 mM Tris-HCl pH 8
1 mM EDTA
0.01% SDS
Filter sterilize
1x protease inhibitor
0.2 mM PMSF
RIPA 300 buffer
50 mM Tris-HCl pH 8
1% NP-40
2 mM EDTA
0.1% Sodium deoxycholate
0.1% SDS
300 mM NaCl
Filter sterilize
RIPA 150 buffer
50 mM Tris-HCl pH 8
1% NP-40
2 mM EDTA
0.1% Sodium deoxycholate
0.1% SDS
150 mM NaCl
Filter sterilize
1x protease inhibitor
0.2 mM PMSF
LiCl/TE wash buffer
0.25 M LiCl
1% NP-40
1% Sodium deoxycholate
10 mM Tris-HCl pH 8
1 mM EDTA pH 8
Sodium bicarbonate elution buffer
1% SDS
0.1 M Sodium bicarbonate
3M NaOAc (Sodium acetate)
20.4g Sodium acetate
to 50 ml ddH2O
Adjust pH to 5.2 with glacial Acetic acid
Other Buffers
Hyalin Extraction Media (for 1 liter)
NaCl 18.5 gm
KCl 0.7 gm
MgSO4 −7H2O 11.9 gm
Glycine 22.5 gm
Tris base 1.21 gm
EGTA 0.76 gm
pH to 8.0
Calcium-free Seawater (for 1 liter)
NaCl 26.5 gm
KCl 0.7 gm
MgSO4 −7H2O 11.9 gm
NaHCO3 0.5 gm
pH to 8.0, salinity should be ~34ppt
Conclusions
Remarkably sensitive and creative new technologies allow us to see more focused and detailed aspects of gene regulation. As new and rapidly changing as these approaches are, the technologies are robust and reproducible. The three approaches we document here in concert allow greater confidence and insight into how a gene regulatory network functions and how it may have evolved. We do end though with the thought these protocols and experimental approaches are only as good as what they tell us about developmental mechanisms. Left to our creativity is how we can incorporate these results into a dynamic, living, and ever changing embryo.
Figure 2:
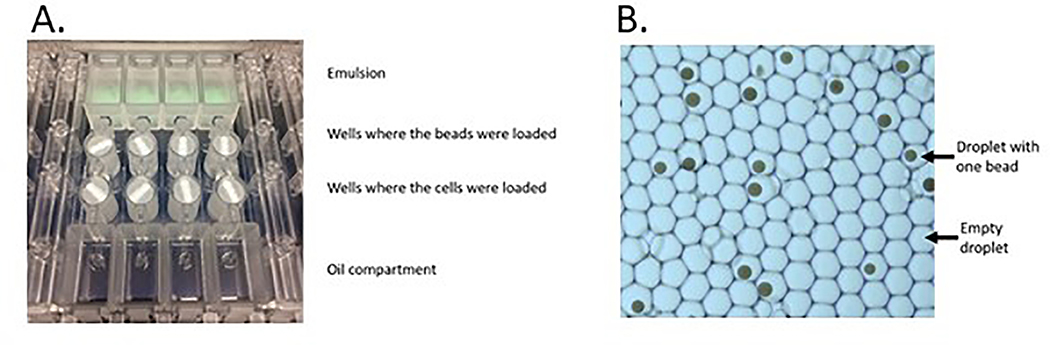
A) The Nadia 4-sample chip. B) The emulsion following a run with oil droplets and beads apparent. The cells have already been lysed and not apparent following a run.
Table 1.
Comparison of single cell sequencing platforms and costs (modified from (Ziegenhain et al., 2017))
| Method | Cell Number per Group* | Minimal Cost ($) |
|---|---|---|
| CEL-seq2/C1 | 85/100/110 | ~2,420/2,310/2,250 |
| Drop-seq | 99/135/254 | ~1,010/700/690 |
| MARS-seq | 110/135/160 | ~1,380/1,030/820 |
| SCRB-seq | 64/90/166 | ~900/810/1,080 |
| Smart-seq/C1 | 150/172/215 | ~9,010/9,440/11,290 |
| Smart-seq2 (commercial Tn5) | 95/105/128 | ~10,470/11,040/13,160 |
| Smart-seq2 (in-house Tn5) | 95/105/128 | ~1,520/1,160/1,090 |
Cell groups listed for sequencing depth of 1, 0.5, and 0.25 million reads.
References
- Barsi JC, Tu Q, Calestani C, & Davidson EH (2015). Genome-wide assessment of differential effector gene use in embryogenesis. Development, 142(22), 3892–3901. doi: 10.1242/dev.127746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp JG, Wollny D, & Treutlein B (2018). Single-cell genomics to guide human stem cell and tissue engineering. Nat Methods, 15(9), 661–667. doi: 10.1038/s41592-018-0113-0 [DOI] [PubMed] [Google Scholar]
- Dal Molin A, & Di Camillo B (2018). How to design a single-cell RNA-sequencing experiment: pitfalls, challenges and perspectives. Brief Bioinform. doi: 10.1093/bib/bby007 [DOI] [PubMed] [Google Scholar]
- Davidson E (1976). Gene Activity in Early Development. New York: Academic Press. [Google Scholar]
- Fletcher RB, Das D, & Ngai J (2018). Creating Lineage Trajectory Maps Via Integration of Single-Cell RNA-Sequencing and Lineage Tracing: Integrating transgenic lineage tracing and single-cell RNA-sequencing is a robust approach for mapping developmental lineage trajectories and cell fate changes. Bioessays, 40(8), e1800056. doi: 10.1002/bies.201800056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresques TM, & Wessel GM (2018). Nodal induces sequential restriction of germ cell factors during primordial germ cell specification. Development, 145(2). doi: 10.1242/dev.155663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths JA, Scialdone A, & Marioni JC (2018). Using single-cell genomics to understand developmental processes and cell fate decisions. Mol Syst Biol, 14(4), e8046. doi: 10.15252/msb.20178046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque A, Engel J, Teichmann SA, & Lonnberg T (2017). A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med, 9(1), 75. doi: 10.1186/s13073-017-0467-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks SC, Townes FW, Teng M, & Irizarry RA (2018). Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics, 19(4), 562–578. doi: 10.1093/biostatistics/kxx053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, … Linnarsson S (2014). Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods, 11(2), 163–166. doi: 10.1038/nmeth.2772 [DOI] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, … Kuroda MI (2007). MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell, 28(1), 121–133. doi: 10.1016/j.molcel.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, … McCarroll SA (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell, 161(5), 1202–1214. doi: 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClay DR (2004). Methods for embryo dissociation and analysis of cell adhesion. Methods Cell Biol, 74, 311–329. [DOI] [PubMed] [Google Scholar]
- Papalexi E, & Satija R (2018). Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol, 18(1), 35–45. doi: 10.1038/nri.2017.76 [DOI] [PubMed] [Google Scholar]
- Poirion OB, Zhu X, Ching T, & Garmire L (2016). Single-Cell Transcriptomics Bioinformatics and Computational Challenges. Front Genet, 7, 163. doi: 10.3389/fgene.2016.00163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle O, Teichmann SA, & Marioni JC (2015). Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet, 16(3), 133–145. doi: 10.1038/nrg3833 [DOI] [PubMed] [Google Scholar]
- Swartz SZ, Reich AM, Oulhen N, Raz T, Milos PM, Campanale JP, … Wessel GM (2014). Deadenylase depletion protects inherited mRNAs in primordial germ cells. Development, 141(16), 3134–3142. doi: 10.1242/dev.110395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, … Enard W (2017). Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell, 65(4), 631–643 e634. doi: 10.1016/j.molcel.2017.01.023 [DOI] [PubMed] [Google Scholar]