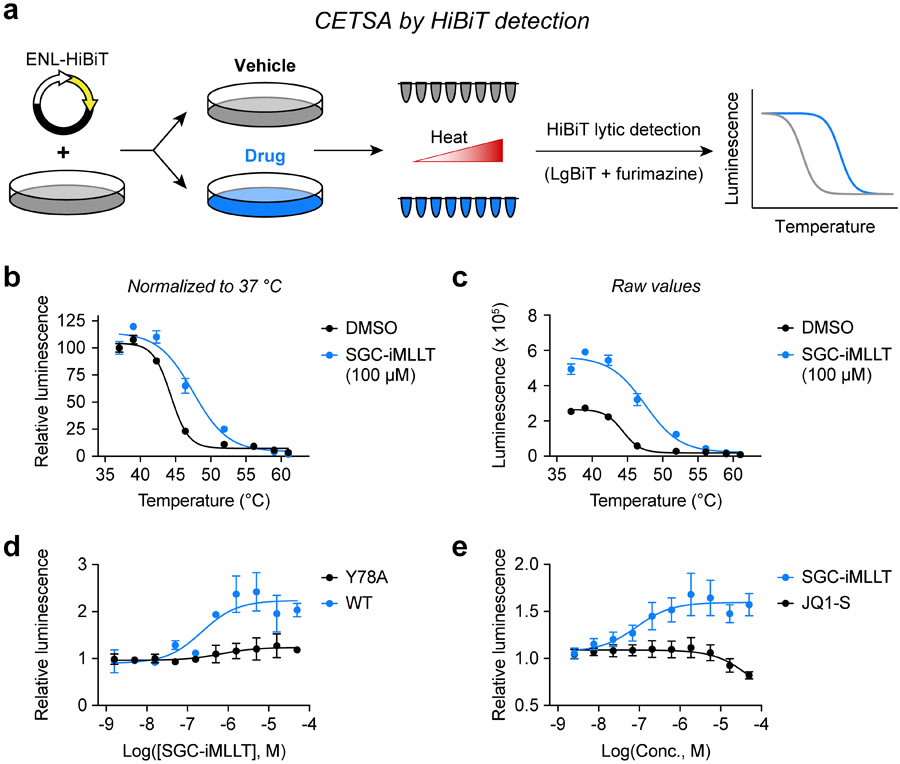
Figure 1.
Discovery of a simplified CETSA protocol for YEATS domains. (a) Schematic depiction of CETSA melt curves using the HiBiT tag to enable detection of soluble protein from homogenous cell lysates. (b) CETSA melt curve of ENL(YEATS)-HiBiT in HEK293T cells after 1-hour treatments with drug or vehicle (N = 4, mean ± standard error of the mean, s.e.m.). Luminescence is normalized to 37 °C. (c) Raw luminescence values from the experiment depicted in (b). (d) ITDR at 37 °C for HEK293T cells expressing wild-type or mutant ENL(YEATS)-HiBiT and treated for 1 hour with SGC-iMLLT (N = 2, mean ± s.e.m.). Luminescence is normalized to DMSO control. (e) ITDR at 37 °C for OCI/AML-2 cells stably expressing AF9(YEATS)-HiBiT and treated for 1 hour (N = 4, mean ± s.e.m.). Luminescence is normalized to DMSO control.