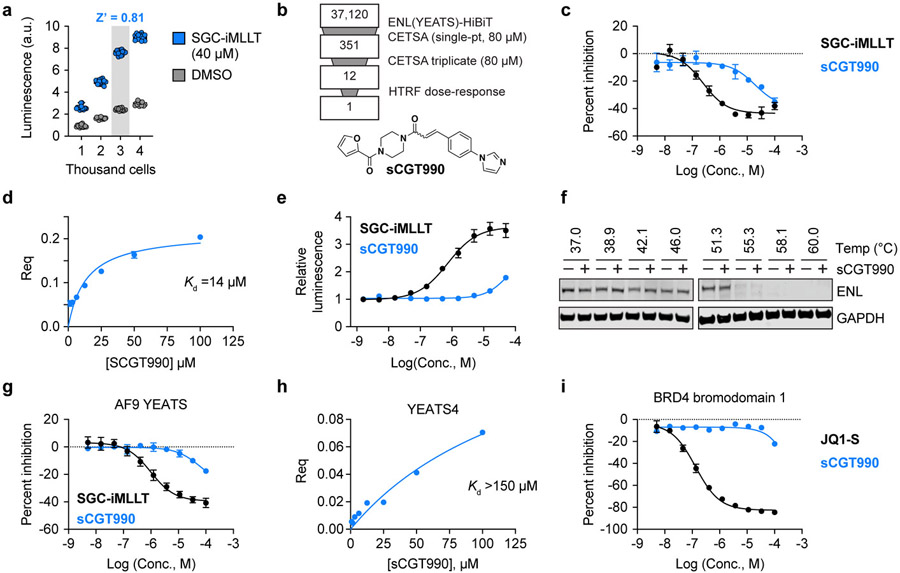
Figure 3.
High-throughput ligand discovery for ENL YEATS in living cells. (a) Optimization of cell density for 1536-well formatted assay. Luminescence values are shown for vehicle and drug-treated OCI/AML-2 cells stably expressing ENL(YEATS)-HiBiT at increasing cell concentrations (N = 15). (b) Summary of high-throughput screen to identify ENL YEATS domain inhibitors. Chemical structure of sCGT990 is depicted at bottom. (c) Dose-dependent inhibition of homogenous time-resolved FRET (HTRF) signal generated by ENL YEATS association with H3K27cr. SGC-iMLLT is included as a positive control (N = 2, mean ± s.e.m.). (d) Biolayer interferometry validation of sCGT990 binding to immobilized biotin-ENL(YEATS). (e) Dose-dependent stabilization of ENL(YEATS)-HiBiT by sCGT990 and positive control compound, SGC-iMLLT. Luminescence values are depicted relative to DMSO (N = 3, mean ± s.e.m.). (f) Immunoblot analysis of CETSA melt curve for endogenous ENL protein in OCI/AML-2 cells treated with sCGT990 (80 μM) or vehicle (DMSO) for 1 hr. (g) Dose-dependent inhibition of HTRF signal generated by AF9 YEATS association with H3K27cr. SGC-iMLLT is included as a positive control (N = 2, mean ± s.e.m.). (h) Biolayer interferometry measures of sCGT990 binding to immobilized biotin-YEATS4(YEATS). (i) Same as in (g), but for BRD4 bromodomain 1 and tetra-acetyl H4 (JQ1-S included as a positive control, (N = 2, mean ± s.e.m.).