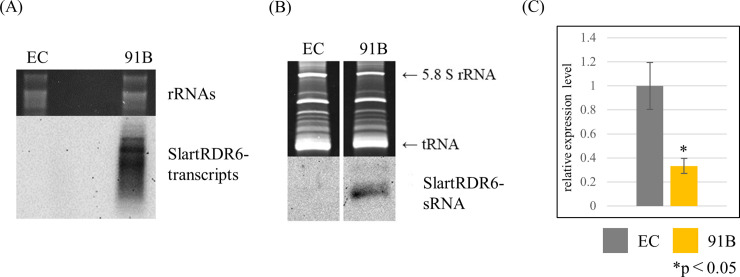
Fig 1. Evaluation of SlRDR6 suppression efficiency.
Transgenic tomato plants were generated to analyze a role of SlRDR6 upon PSTVd infection. (A) SlartRDR6-transcripts and (B) SlartRDR6-sRNA were detected by Northern-blot hybridization with DIG-labeled cRNA probe for SlartRDR6. rRNAs and tRNA were stained with ethidium bromide and used as a loading control. SlartRDR6-transcripts and SlartRDR6-sRNA were only detected in line 91B. (C) Expression levels of endogenous SlRDR6 mRNA were analyzed by RT-qPCR performed with the PCR primers for the endogenous SlRDR6 gene. Mean values are based on three biological replicates of the pooled sample which is collected from five individual plants. The relative expression levels were calculated with the value of EC plants as a standard. The expression level of endogenous SlRDR6 mRNA in line 91B was significantly reduced to approximately 40% of that in EC plants. The significant decrease of SlRDR6 expression level in line 91B was confirmed by Welch’s t-test.