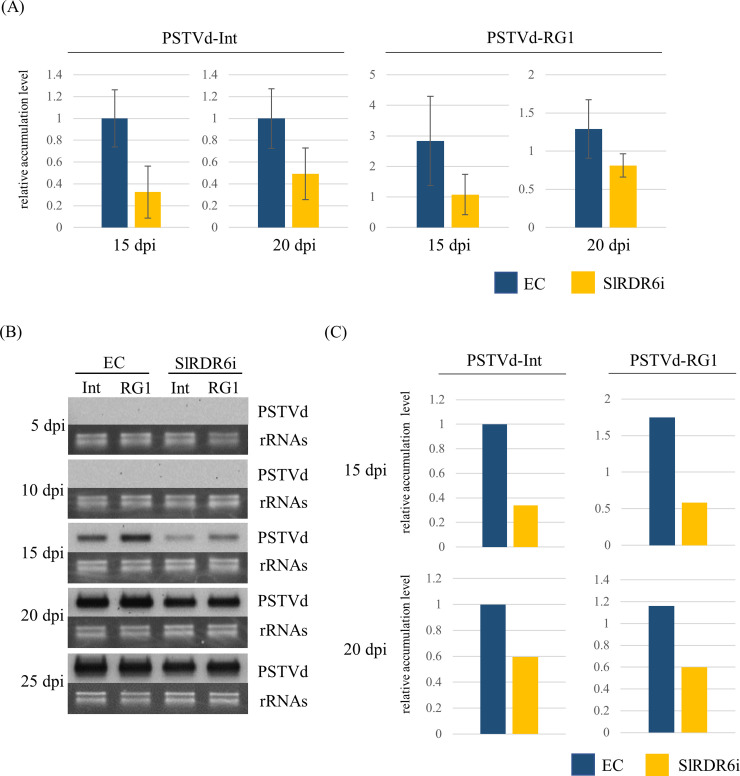
Fig 4. Time-course analysis of accumulation levels of PSTVd genomic RNA in transgenic ‘Moneymaker’ tomato plants.
(A) Accumulation of PSTVd genomic RNA was analyzed by RT-qPCR. At 15 and 20 dpi, the accumulation levels of PSTVd-Int and PSTVd-RG1 tended to be lower in SlRDR6i than in EC plants. (B) Accumulation of PSTVd genomic RNA was analyzed by Northern-blot hybridization with DIG-labeled cRNA probe for PSTVd. rRNAs were stained with ethidium bromide and used as a loading control. At 15 and 20 dpi, the accumulation levels of PSTVd-Int and PSTVd-RG1 were lower in SlRDR6i than in EC plants. (C) Comparison of relative accumulation of PSTVd genomic RNA by Northern-blot hybridization. Signals of Northern-blot hybridization indicating the accumulation of PSTVd genomic RNA were quantified using Quantity One (version 4.6.2) software package. The relative PSTVd levels were calculated for each time point with the value of EC plants inoculated with PSTVd-Int as a standard. The accumulation of the two PSTVd strains in SlRDR6i plants at 15 dpi reached about one-third of that in EC plants. qPCR was performed with the PCR primer for PSTVd. Mean values are based on three biological replicates of the pooled sample which is collected from five individual plants.