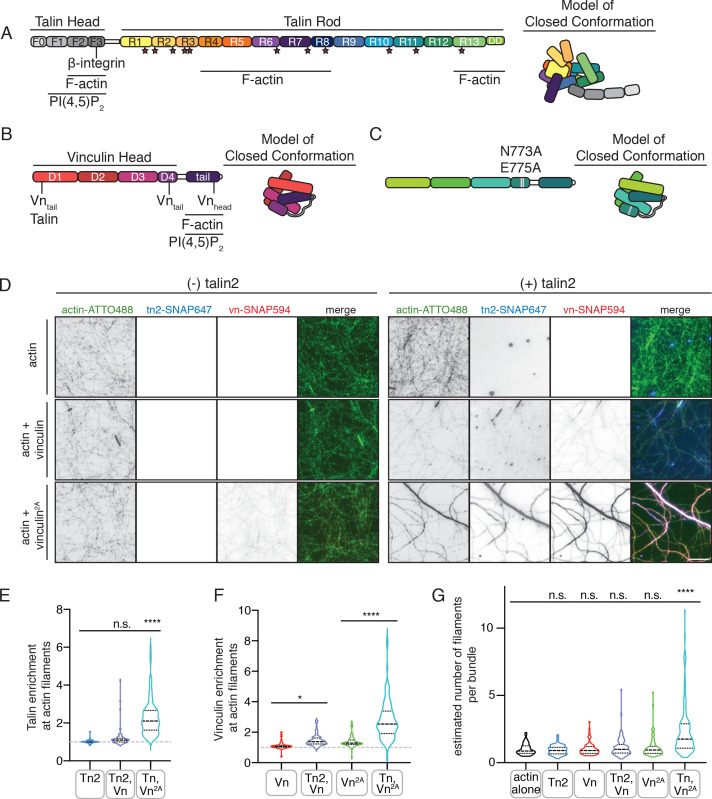
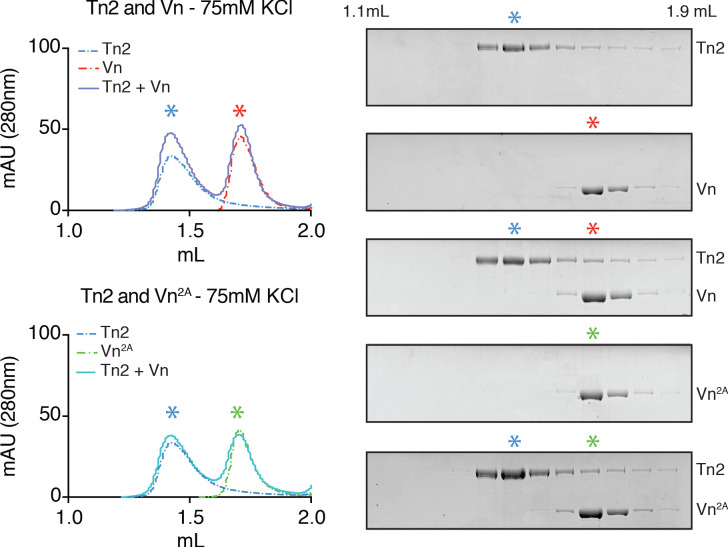
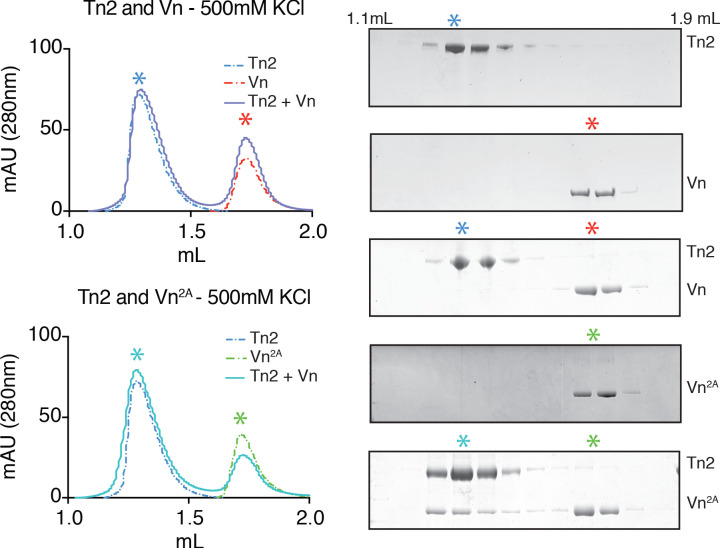
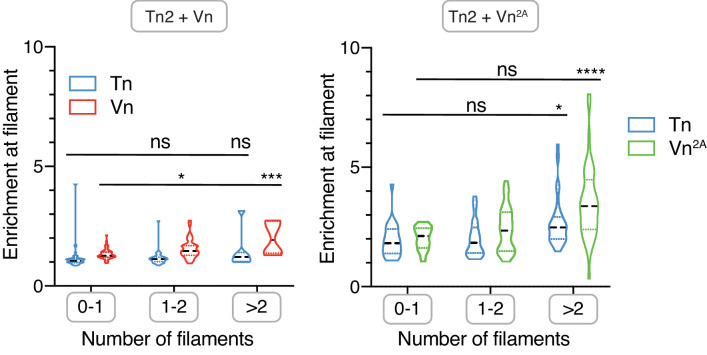
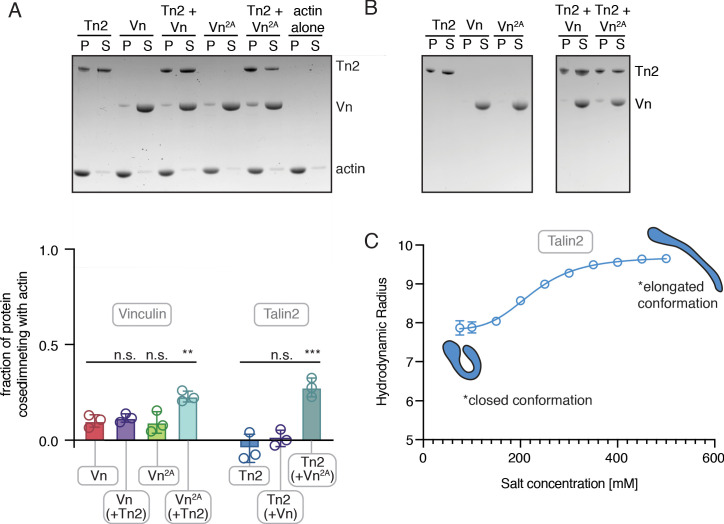
Figure 1. Autoinhibition blocks interactions between talin, vinculin, and actin in vitro.
(A) Human talin2 domain organization, left. Stars highlight predicted vinculin binding sites. To the right, a model of the closed, autoinhibited conformation of talin, based on the Tn1 autoinhibited structure (Dedden et al., 2019). (B) Human vinculin domain organization, with model of autoinhibited structure (red) and (C) human vinculin with a deregulating mutation in the D4 domain (N773A,E775A), with a model of the partially deregulated structure (green) (Bakolitsa et al., 2004; Cohen et al., 2005). (D) Representative three-color TIRF microscopy images of 1.5 µM SNAP-tag-labeled FA proteins and 1 µM actin (10% actin-ATTO488,green) added to TIRFm buffer (10 mM imidazole, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.2 mM ATP, pH 7.5) supplemented with 15 mM glucose, 20 µg/mL catalase, 100 µg/mL glucose oxidase, 1 mM DTT and 0.25% methyl-cellulose (4000 cp). Images acquired after 30 min of polymerization. Scale bar = 5 µm. (E) Quantification of talin enrichment at actin filaments, compared to background Tn-SNAP647 signal, for Tn alone, with Vn wild-type, and with Vn2A. A value of 1 indicates no enrichment at filaments. (F) Quantification of vinculin enrichment at actin filaments, compared to background Vn-SNAP647 or Vn2A-SNAP647 signal, both with or without Tn2. A value of 1 indicates no enrichment at filaments. (G) Estimation of number of filaments per actin bundle for each condition shown in (D), n (from left to right)=43, 57, 60, 93, 97, 108. The average fluorescence of individual filaments in actin alone samples was used to define the signal of a single actin filament, and then used to estimate the number of filaments for each condition. n.s. >0.5,*p<0.05 ****p<0.0001, by one-way ANOVA.