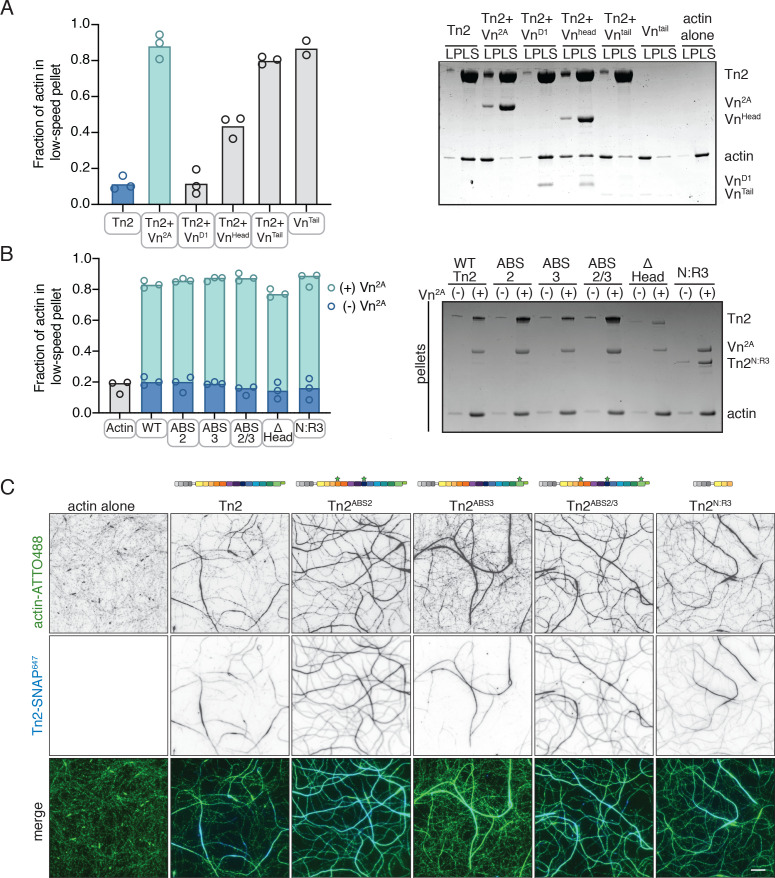
Figure 3. Talin mediates interactions between full-length vinculin and actin.
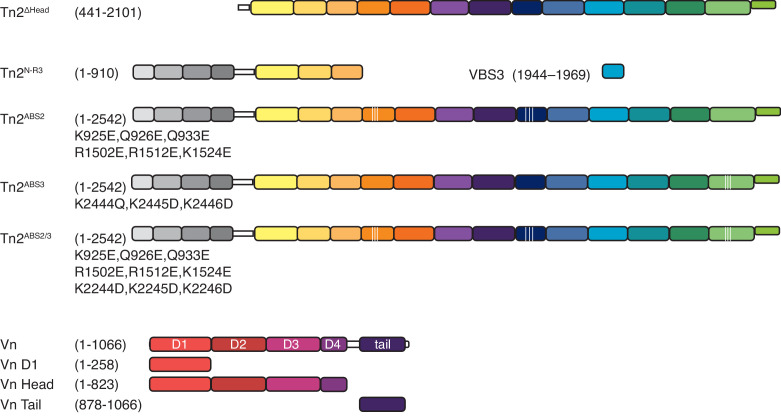
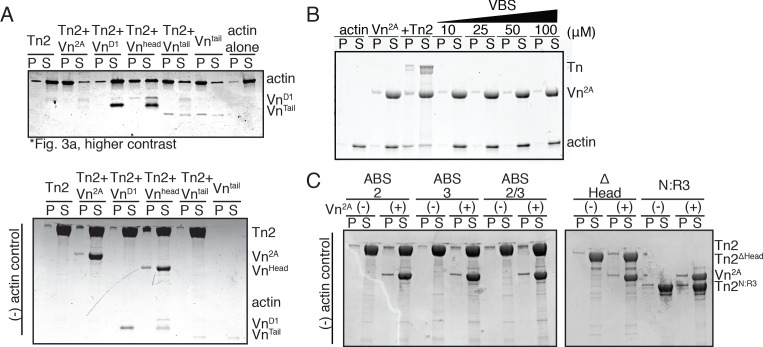
(A) Actin bundling co-sedimentation assay with filamentous actin (2.5 µM) and Tn2 with various vinculin fragments (2.5 µM, see Figure 3—figure supplement 1), supernatant and pellet samples were analyzed using SDS-PAGE. Graph shows individual data points and mean densitometry for three independent samples. (B) Actin bundling co-sedimentation assay with filamentous actin (2.5 µM) and various talin2 mutants and fragments (see Figure 3—figure supplement 1), with and without Vn2A (2.5 µM), supernatant and pellet samples were analyzed using SDS-PAGE. Graph shows individual data points and mean densitometry for talin2 proteins alone (dark blue) and with Vn2A (aqua) for three independent samples. Cosedimentation experiments performed in 20 mM HEPES, pH 7.5, 75 mM KCl, 2 mM MgCl2, and 0.2 mM ATP. Controls for (A,B) found in Figure 3—figure supplement 2.(C) Representative images of two-color TIRFm with 1 µM actin (5% ATTO488-actin) and Vn2A (1.5 µM) with different SNAP-647-labeled Tn2 proteins (1.5 µM). Schematics above images indicate domain locations of mutations and truncations. Quantification can be found in Figure 3—figure supplement 4. Scale bar = 5 µm. TIRFm experiments were carried out in TIRFm buffer with 15 mM glucose, 20 µg/mL catalase, 100 µg/mL glucose oxidase, 1 mM DTT and 0.25% methyl-cellulose (4000 cp).