Figure 1. A chemogenomic view of the response to DNA damage.
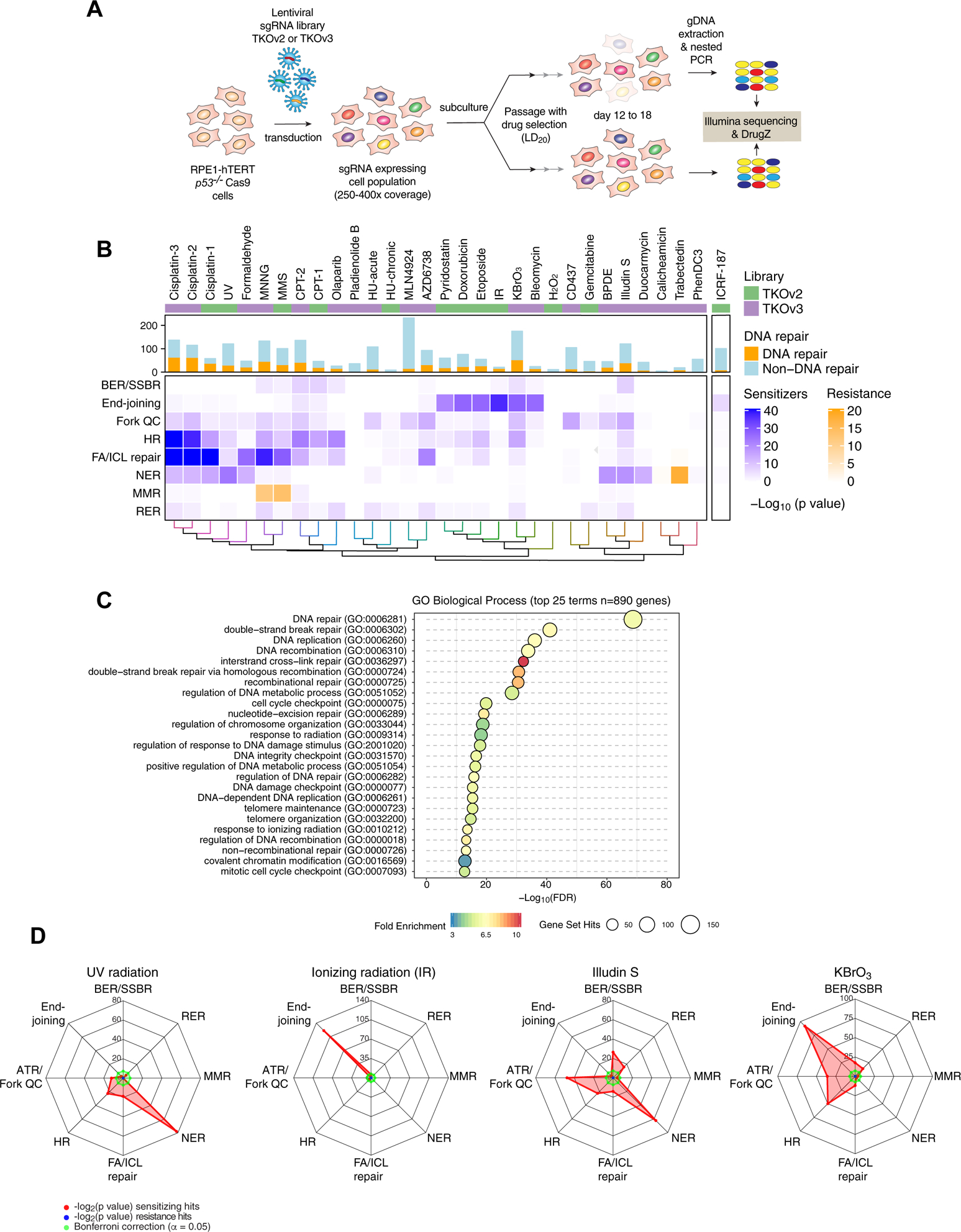
(A) Schematic of the dropout screens.
(B) Heat map representation of the 31 CRISPR screens undertaken in RPE1 hTERT p53−/− Cas9 cells. The histogram indicates the number of hits in each screen and the number of DNA repair factors identified. The lower heat map panel shows the log10-transformed p values for enrichment of different DNA repair pathways (rows) calculated using a one-sided Fisher’s exact test. The color scale indicates fold-enrichment for resistance (orange) and sensitization (blue). Pathways are defined in the text except for ribonucleotide excision repair (RER).
(C) Top 25 enriched GO terms, biological process, identified using g:Profiler (>10-fold enrichment; p < 0.05, with Benjamini-Hochberg FDR correction) among all 890 hits.
(D) Radar plots of the indicated genotoxic CRISPR screens depicting sensitization (red) or resistance (blue) for different DNA repair pathways. Values indicate the log2-transformed p values of the Fisher’s exact test score. Bonferroni thresholds are in green.
See also Figure S1.
