Figure 4. ERCC6L2 promotes canonical NHEJ.
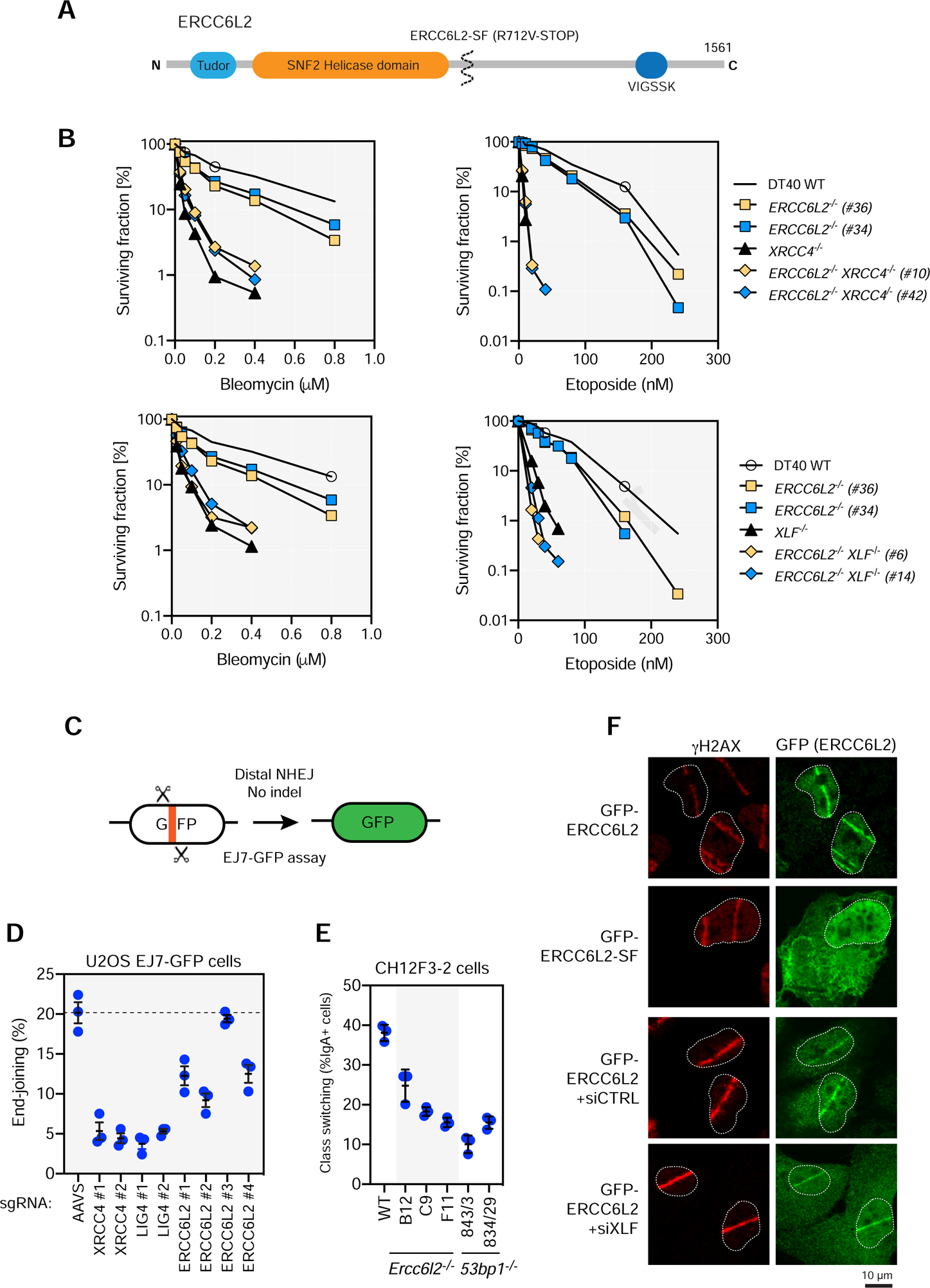
(A) Schematic overview of ERCC6L2. ERCC6L2 also possess a short isoform of 712 residues produced by alternative splicing. Most disease-associated ERCC6L2 alleles produce proteins that are truncated prior to residue 712.
(B) Cell proliferation assays of DT40 cells of the indicated genotypes treated with either etoposide or bleomycin for 3 d. Note that the bleomycin proliferation assays were done as part of the same experiment but were separated for clarity. Data presented as the mean of a technical triplicate. An independent experiment is shown in Figure S5C.
(C) Schematic of the EJ7-GFP NHEJ assay.
(D) End-joining frequency of U2OS-EJ7 cells following depletion of the indicated genes by gene editing. Dashed line represents the mean of the end-joining frequency of the AAVS1-targeted condition. Data presented as mean ± SD; N=3. TIDE analysis is shown in Table S6.
(E) Class switch recombination levels (% IgA+ cells) in CH12F3–2 cells of the indicated genotypes. Data presented as mean ± SD; N=3.
(F) U2OS cells transfected with a plasmid encoding either full-length GFP-ERCC6L2 or the short isoform (SF) were subjected to laser microirradiation and then processed for immunofluorescence with GFP and γH2AX antibodies 30 min post-irradiation. The mean percentage of cells (± SD; N=3) with γH2AX- and GFP+ stripes are indicated.
See also Figure S5.
