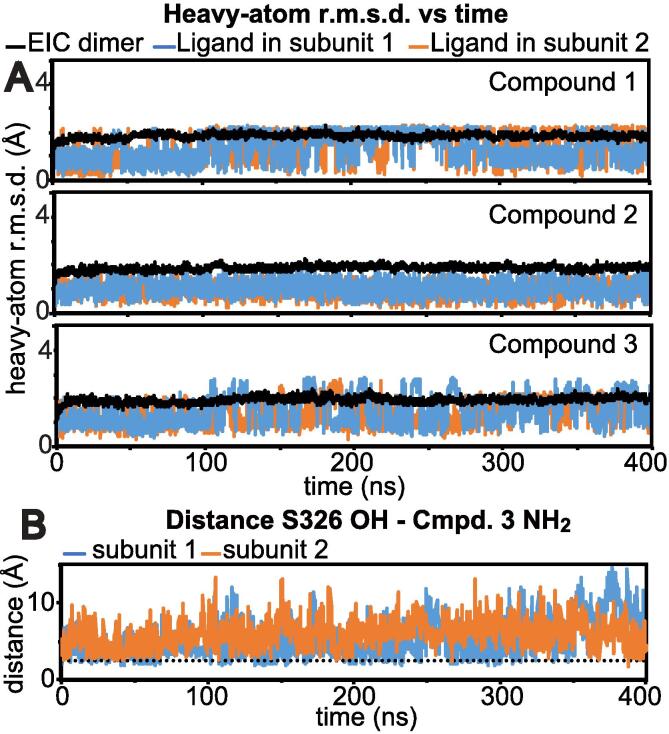
Fig. 5.
MD simulations of the EIC-inhibitor complexes. (a) Heavy-atom r.m.s.d. to the docking structure versus time calculated for the 400-ns MD run on the EIC-inhibitor complexes. EIC was simulated in its physiological dimeric form with inhibitors bound to both subunits. The r.m.s.d. calculated for the EIC, the inhibitor bound to the first subunit, and the inhibitor bound to the second subunit are colored black, light blue, and orange, respectively. Top, center, and bottom plots are for the complexes with compound 1, 2, and 3, respectively. (b) The distance between the hydroxyl group of S326 and the amine group of compound 3 is plotted versus time. Data for subunit 1 and 2 are colored light blue and orange, respectively. The dotted line is at 2.4 Å to indicate the distance required for hydrogen-bond formation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)