Abstract
Background & Aims
Crohn’s disease (CD) likely represents decreased immune tolerance to intestinal bacterial antigens. Most CD patients have high titers of antibodies to intestinal commensal proteins, including the outer membrane porin C (OmpC) of Escherichia coli.
Methods
By using major histocompatibility complex II tetramers, we identified an HLA-DRB1∗15:01-restricted peptide epitope of OmpC recognized by CD4+ T cells in peripheral blood mononuclear cells from HLA-DRB1∗15:01+ healthy control (HC) and CD patients.
Results
The precursor frequency of these cells in CD correlated with anti-OmpC IgA titers, but did not differ from that of HCs. In both cohorts, they showed a CD161+, integrin α4β7+ phenotype ex vivo by flow cytometry, distinct from the C-X-C Motif Chemokine Receptor 3 phenotype of autologous influenza hemagglutinin (Flu) peptide-specific T cells. The T-cell receptor α and β chains of in vitro–expanded OmpC-specific T-cell clones often contained public amino acid sequences that were identical in cells from different patients. Expanded T-cell clones from CD subjects produced significantly less interleukin (IL)10 (P < .0001) than those from HCs, and a trend toward decreased production of the T helper 2 cell–associated IL4, IL5, and IL13 by CD clones also was seen.
Conclusions
Both HCs and CD patients have detectable OmpC-specific T cells in circulation, with similar immunophenotypes and often identical T-cell–receptor sequences. However, expanded clones from patients with CD produce less of the immunoregulatory cytokine IL10, showing a selective defect in the regulatory function of intestinal microbial antigen-specific T cells in patients with CD.
Keywords: Public TCR, OmpC, IL10, Tetramer-Guided Epitope Mapping
Abbreviations used in this paper: CD, Crohn’s disease; cDNA, complementary DNA; CXCR3, C-X-C Motif Chemokine Receptor 3; Fc, fragment crystallizable region; FITC, fluorescein isothiocyanate; Flu, influenza hemagglutinin; HC, healthy control; IBD, inflammatory bowel disease; IFN, interferon; IL, interleukin; MHC, major histocompatibility complex; OmpC, outer membrane porin C; PE, phycoerythrin; PerCP, peridinin-chlorophyll-protein complex; TCR, T-cell receptor; TGEM, tetramer-guided epitope mapping; Th, helper T cell; TIGIT, T cell immunoreceptor with Ig and ITIM domains
Graphical abstract
Synopsis.
We identified and characterized blood T cells specific for a ubiquitous gut flora antigen (outer membrane porin C protein of Escherichia coli), to which Crohn’s disease patients often make antibodies. These showed a Crohn’s-specific defect in immunoregulatory cytokine (interleukin 10) production, as well as public T-cell receptors.
Crohn’s disease (CD) is a chronic, relapsing-and-remitting inflammatory disease of the intestinal mucosa. Although incompletely understood, immune dysregulation with a loss of tolerance to intestinal bacterial antigens has been implicated in the pathogenesis of CD. Antibodies against the commensal flora identified with high incidence in the serum of patients with CD but not healthy controls (HCs) include outer membrane porin C (OmpC) of Escherichia coli, mannose epitopes from the yeast Saccharomyces cerevisiae, and bacterial flagellin proteins (CBir).1,2 The serum titers of such antibodies are correlated not only with the diagnosis of CD, but also with the location and severity of CD lesions.2, 3, 4, 5 Although the antibodies themselves are not thought to be pathogenic, they represent concrete evidence of immune dysregulation and loss of tolerance to commensal gut flora in inflammatory bowel disease (IBD).1
Although microbe-specific antibodies are well described, little is known about gut flora-specific cellular immunity in IBD. Because B cells typically require help from CD4+ T cells recognizing peptides from the same antigen to produce circulating high-affinity antibodies,6 it is likely that T cells recognizing these antigens are present in patients with CD. In mice, it has been shown that CBir1-specific immunoglobulin production is T-cell dependent.7 In fact, major histocompatibility complex (MHC) class II–dependent CBir1-specific T-cell responses have been shown from peripheral blood mononuclear cells (PBMCs) and intestinal lymphocytes in patients with CD.8 Interestingly, although the study may have been underpowered, there was no correlation between anti-CBir1 titers and CBir1-directed T-cell responses.8 More recently, polyclonal interferon (IFN)-γ8 and interleukin (IL)17A-producing T cells9 that are responsive to intact bacterial flagellin proteins have been described in Crohn’s patients, albeit at precursor frequencies high enough (up to 20% of total CD4+ T cells) to suggest antigen-nonspecific or bystander activation. Flagellin itself is an agonist for the innate immune receptor Toll-like receptor 5,10 and thus may have atypical properties as a commensal antigen.
We therefore used a more reductionist approach, using MHC II tetramer-guided epitope mapping (TGEM)11 to identify a single peptide epitope from the bacterial antigen OmpC recognized by HLA-DRB1∗15:01-restricted T cells. We then were able to identify, quantify, characterize, and isolate individual HLA-DRB1∗15:01-restricted T cells specific for this OmpC peptide from the peripheral blood. We found OmpC-specific T cells to be present in both HCs and CD patients. This allowed us to compare the immunophenotype of OmpC-specific T cells between HC and CD patients, and between OmpC-specific T cells and autologous influenza peptide-specific T cells. OmpC-specific T-cell clones were expanded in vitro for functional analyses, which showed clear differences in antigen-specific cytokine production between clones from CD patients and HCs.
Results
Identification of a Peptide Epitope of OmpC Recognized by T Cells
HLA-DRB1 allele frequencies of CD patients in our cohort who were seropositive for either OmpC or the flagellin protein, CBir1 (Table 1), were compared with allele frequencies of Caucasians in the United States (from http://www.allelefrequencies.net) (Figure 1). Almost half of OmpC-seropositive patients were HLA-DRB1∗15:01-positive in at least 1 allele, which is roughly twice the allele frequency in either the general public or in our CBir1-seropositive CD patients cohort (Figure 1). This suggested that an HLA-DRB1∗15:01-restricted T-cell antigen in the peptide sequence of OmpC may drive anti-OmpC antibody formation.
Table 1.
HLA Genotype vs Serology
| Patient, n | OmpC EU | CBir1 EU | Positive serology | DRB1(a)a | DRB1(b)a | DQB1(a)a | DQB1(b)a | SSP DRB 1(a)a low res | SSP DRB 1(b)a low res | SSP (a)a high res | SSP (b)a high res | Final DRB 1(a)a | Final DRB 1(b)a |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 45 | 45 | OmpC + cBir | (∗1501) | 01 | 15 | 0103 | ND | 0103 | 1501 | |||
| 2 | 51 | 13 | OmpC | (∗0401) | (∗1501) | 04 | 15 | ND | 1501 | 0401 | 1501 | ||
| 3 | 45 | 7 | OmpC | (∗1501) | 14 | 15 | 1501 | 14x | 1501 | ||||
| 4 | 32 | 11 | OmpC | (∗03) | (∗03) | 03 | 0301 | 0301 | 0301 | 0301 | |||
| 5 | 61 | 16 | OmpC | (∗1501) | 15 | 1501 | 1501 | ||||||
| 6 | 14 | 31 | cBir1 | (∗0401) | (∗0401) | 0401 | 0401 | 0401 | 0401 | ||||
| 7 | 27 | 19 | OmpC | (∗1501) | 15 | 07 | 1501 | 1501 | 07x | ||||
| 8 | 55 | 32 | OmpC + cBir | (∗13) | 13 | 13 | ND | ND | 13x | 13x | |||
| 9 | 31 | 17 | OmpC | (∗0401) | (∗01) | 04 | 01 | ND | 0101 | 0401 | 0101 | ||
| 10 | 9 | 26 | cBir1 | (∗1501) | 15 | 1501 | 1501 | 1501 | 1501 | ||||
| 11 | 25 | 56 | OmpC + cBir | (∗0401) | (∗1501) | (∗0602) | 15 | 04 | ND | 1501 | 0401 | 1501 | |
| 12 | 56 | 7 | OmpC | (∗0401) | (∗03) | 04 | 0301 | ND | 0401 | 0301 | |||
| 13 | 64 | 36 | OmpC + cBir | (∗1501) | (∗0602) | 08 | 15 | 1501 | 08x | 1501 | |||
| 14 | 10 | 49 | cBir1 | (∗13) | 04? | 13 | 0103 | ND | 0103 | 13x | |||
| 15 | 24 | 42 | OmpC + cBir | (∗13) | (∗1501) | 13 | 15 | ND | 1501 | 13x | 1501 | ||
| 16 | 10 | 37 | cBir1 | (∗01) | 01 | 0101 | 0101 | 0101 | |||||
| 17 | 14 | 27 | cBir1 | (∗0405) | (∗13) | 04 | 13 | ND | ND | 0405 | 13x | ||
| 18 | 34 | 9 | OmpC | 11 | 13 | ND | 11x | 13x | |||||
| 19 | 17 | 38 | cBir1 | (∗03) | (∗02) | 01 | 03 | 0103 | 0301 | 0103 | 0301 | ||
| 20 | 20 | 33 | cBir1 | (∗13) | 13 | 07? | ND | 1001 | 1001 | 13x | |||
| 21 | 9 | 143 | cBir1 | (∗0401) | (∗13) | 04 | 13 | ND | ND | 0401 | 13x | ||
| 22 | 38 | 19 | OmpC | (∗03) | 03 | 14 | 0301 | 1401 | 0301 | 1401 | |||
| 23 | 8 | 30 | cBir1 | (∗13) | 07 | 13 | ND | 07x | 13x | ||||
| 24 | 8 | 31 | cBir1 | (∗0401) | 04 | 0401 | 0401 | 0401 | |||||
| 25 | 34 | 36 | OmpC + cBir | (∗0401) | 04 | 0401 | 0401 | 0401 | |||||
| 26 | 8 | 118 | cBir1 | (∗01) | (∗13) | 01 | 13 | 0101 | ND | 0101 | 13x | ||
| 27 | 6 | 26 | cBir1 | 07 | 14 | Contam | ND | x | 14x | ||||
| 28 | 11 | 46 | cBir1 | (∗0402) | 04 | 08 | ND | ND | 0402 | 08x | |||
| 29 | 7 | 42 | cBir1 | (∗0401) | (∗1501) | (∗0602) | (∗0302) | 04 | 15 | ND | 1501 | 0401 | 1501 |
| 30 | 20 | 32 | cBir1 | 07 | 11x | ||||||||
| 31 | 30 | 26 | OmpC + cBir | (∗03) | (∗1501) | 03 | 15 | 0301 | ND | 0301 | 1501 | ||
| 32 | 25 | 26 | OmpC + cBir | 11 | 11 | 11x | 11x | ||||||
| 33 | 21 | 40 | cBir1 | (∗02) | 07 | 11 | 0701 | ND | 0701 | 11x | |||
| 34 | 12 | 49 | cBir1 | (∗01) | (∗0401) | (∗0302) | 01 | 04 | 0101 | ND | 0101 | 0401 | |
| 35 | 47 | 20 | OmpC | 08 | 11 | 08x | 11x | ||||||
| 36 | 30 | 12 | OmpC | (∗0404) | 04 | 07 | ND | 0404 | 11x |
EU, enzyme-linked immunosorbance assay (ELISA) units; res, resolution; SSP, sequence specific primer.
Lowercase numbers in parentheses are alleles of the indicated HLA locus.
Figure 1.
The HLA-DRB1∗15:01 genotype is enriched among OmpC-seropositive Crohn’s patients. DNA from CD patients with known seropositivity to OmpC (n = 19) or cBir1 (n = 25) was genotyped for HLA-DRB as described in the Materials and Methods section. For HLA-DRB1 genes in families 07, 08, 11, 13, and 14, subtyping was not performed, and thus is indicated by a “??” suffix. The frequency of each allele in each cohort is shown, alongside the HLA-DRB1 allele frequencies observed in a cohort of 61,655 Caucasians sampled from throughout the United States for reference (Naval Medical Research Centre, and Department of Microbiology and Immunology, Georgetown University, 2002, unpublished data). Asterisks are shown above alleles for the Crohn’s cohorts that showed a significant (P < .05) difference in frequency from this reference population by the chi-squared test with Yates correction.
We therefore used fresh PBMCs from OmpC-seropositive, HLA-DRB1∗15:01+ CD patients to search an OmpC peptide library by TGEM as described previously11 (see Materials and Methods section for TGEM description). Two overlapping peptides (p41, OmpC 321-340, and p42, OmpC 329-348, with the amino acid sequences VGATYYFNKNMSTYVDYKIN and KNMSTYVDYKINLLDDNQFT, respectively) containing a shared amino acid sequence (KNMSTYVDYKIN) near the C-terminus of OmpC were identified as potential T-cell epitopes. These peptides were identified from the E coli reference sequence WP_000865568.1, which lacks the first 16 amino acids of the full-length E coli K-12 reference sequence (GenBank accession number: CQR81715.1). We chose the NCBI reference sequence WP_000865568.1 because it was predicted that the first 16 amino acids of this segment are cleaved intracellularly before the protein reaches the outer surface of the cell. TGEM of these peptides was repeated using the fresh PBMCs from 3 HLA-DRB1∗15:01+ HC donors, all of whom confirmed specificity for OmpC 321-340 and OmpC 329-348 (Figure 2). OmpC 321-340 was used subsequently throughout all experiments to identify OmpC-specific T-cell populations.
Figure 2.
TGEM identifies an HLA-DRB1∗15:01-restricted T-cell antigen from OmpC. (A) PBMCs from an OmpC-seropositive CD HLA-DR 1501 subject were stimulated with 9 pools of OmpC peptides and subsequently stained with the corresponding pooled peptide tetramers. Tetramer staining was observed in pool 9. Stimulated PBMCs from pool 9 then were stained with tetramers containing the 4 individual peptides (p41–p44) of pool 9. (B) PBMCs from 3 HCs were stimulated with 9 pools of OmpC peptides and subsequently stained with the corresponding pooled peptide tetramers. (C) HC PBMCs from pool 9 were stained with its individual peptides (p41–p44). (B and C) The percentage of all CD4+ T cells staining positive for tetramers containing indicated pools or peptides is shown. p41 corresponds to OmpC 321–340. p42 corresponds to OmpC 329–348.
By using NCBI BLAST protein suite (https://blast.ncbi.nlm.nih.gov/Blast.cgi), we found the OmpC 321-340 peptide sequence to align highly with the Enterobacteriaceae family (Table 2). We found 7 E coli organisms to have 100% alignment with our peptide sequence. Avirulent species such as H605 were found, although no Crohn’s-related E coli species such as LF82, CFT073, and UM146 were identified. Other organisms within Enterobacteriaceae such as Klebsiella and Shigella genii also aligned with this peptide sequence. In summary, this peptide sequence appears to be highly conserved across this bacteria family, containing both commensals and pathobionts of the human gut flora.
Table 2.
NCBI Protein BLAST Taxonomy OmpC321–340
| Taxonomy | Hits, n | Organisms, n | Description |
|---|---|---|---|
| Root | 171 | 60 | |
| Bacteria | 170 | 59 | |
| Enterobacterales | 169 | 58 | |
| Enterobacteriaceae | 1 | 57 | Enterobacteriaceae hits |
| Escherichia | 64 | 8 | |
| Escherichia species E4736 | 2 | 1 | Escherichia species E4736 hits |
| Escherichia coli | 56 | 7 | E coli hits |
| E coli HS | 1 | 1 | E coli HS hits |
| E coli 541-1 | 1 | 1 | E coli 541-1 hits |
| E coli H605 | 1 | 1 | E coli H605 hits |
| E coli H736 | 1 | 1 | E coli H736 hits |
| E coli TA464 | 1 | 1 | E coli TA464 hits |
| E coli O43 str. RM10042 | 1 | 1 | E coli O43 str. RM10042 hits |
| Enterobacter cloacae complex | 15 | 4 | |
| Enterobacter hormaechei | 3 | 1 | E hormaechei hits |
| E cloacae | 10 | 2 | E cloacae hits |
| E cloacae BWH 43 | 1 | 1 | E cloacae BWH 43 hits |
| Enterobacter species MGH 10 | 1 | 1 | Enterobacter species MGH 10 hits |
| Klebsiella | 38 | 11 | |
| Klebsiella pneumoniae | 27 | 7 | K pneumoniae hits |
| K pneumoniae subspecies pneumoniae | 1 | 1 | K pneumoniae subspecies pneumoniae hits |
| K pneumoniae UCI 41 | 1 | 1 | K pneumoniae UCI 41 hits |
| K pneumoniae BIDMC 53 | 1 | 1 | K pneumoniae BIDMC 53 hits |
| K pneumoniae BIDMC 46a | 1 | 1 | K pneumoniae BIDMC 46a hits |
| K pneumoniae UHKPC45 | 1 | 1 | K pneumoniae UHKPC45 hits |
| K pneumoniae IS46 | 1 | 1 | K pneumoniae IS46 hits |
| Klebsiella oxytoca | 2 | 1 | K oxytoca hits |
| Klebsiella quasipneumoniae | 1 | 1 | K quasipneumoniae hits |
| Klebsiella variicola | 1 | 1 | K variicola hits |
| Klebsiella michiganensis | 1 | 1 | K michiganensis hits |
| Salmonella | 22 | 9 | |
| Salmonella enterica | 6 | 9 | S enterica hits |
| S enterica subspecies enterica | 13 | 6 | |
| S enterica subspecies enterica serovar Heidelberg str. N1536 | 1 | 1 | S enterica subspecies enterica serovar Heidelberg str. N1536 hits |
| S enterica subspecies enterica serovar Typhimurium | 5 | 1 | S enterica subspecies enterica serovar Typhimurium hits |
| S enterica subspecies enterica serovar Derby | 1 | 1 | S enterica subspecies enterica serovar Derby hits |
| S enterica subspecies enterica serovar Enteritidis | 3 | 1 | S enterica subspecies enterica serovar Enteritidis hits |
| S enterica subspecies enterica serovar Kentucky | 2 | 1 | S enterica subspecies enterica serovar Kentucky hits |
| S enterica subspecies enterica serovar Agona str. 26 F 98 | 1 | 1 | S enterica subspecies enterica serovar Agona str. 26.F.98 hits |
| S enterica subspecies arizonae | 2 | 1 | S enterica subspecies arizonae hits |
| S enterica subspecies diarizonae | 1 | 1 | S enterica subspecies diarizonae hits |
| Shigella | 26 | 22 | |
| Shigella sonnei | 2 | 1 | S sonnei hits |
| Shigella flexneri | 4 | 20 | S flexneri hits |
| S flexneri 2003036 | 1 | 1 | S flexneri 2003036 hits |
| S flexneri Shi06HN006 | 1 | 1 | S flexneri Shi06HN006 hits |
| S flexneri 2a | 1 | 3 | S flexneri 2a hits |
| S flexneri 2a str. 2457T | 1 | 1 | S flexneri 2a str. 2457T hits |
| S flexneri 2a str. 301 | 1 | 1 | S flexneri 2a str. 301 hits |
| S flexneri 4343-70 | 1 | 1 | S flexneri 4343-70 hits |
| S flexneri K-671 | 1 | 1 | S flexneri K-671 hits |
| S flexneri 2930-71 | 1 | 1 | S flexneri 2930-71 hits |
| S flexneri VA-6 | 1 | 1 | S flexneri VA-6 hits |
| S flexneri K-218 | 1 | 1 | S flexneri K-218 hits |
| S flexneri K-272 | 1 | 1 | S flexneri K-272 hits |
| S flexneri K-304 | 1 | 1 | S flexneri K-304 hits |
| S flexneri K-227 | 1 | 1 | S flexneri K-227 hits |
| S flexneri SFJ17B | 1 | 1 | S flexneri SFJ17B hits |
| S flexneri 2850-71 | 1 | 1 | S flexneri 2850-71 hits |
| S flexneri K-1770 | 1 | 1 | S flexneri K-1770 hits |
| S flexneri K-404 | 1 | 1 | S flexneri K-404 hits |
| S flexneri 6603-63 | 1 | 1 | S flexneri 6603-63 hits |
| S flexneri 2b | 1 | 1 | S flexneri 2b hits |
| Shigella boydii | 1 | 1 | S boydii hits |
| Citrobacter species TSA-1 | 1 | 1 | Citrobacter species TSA-1 hits |
| Leclercia adecarboxylata | 1 | 1 | L adecarboxylata hits |
| Edwardsiella tarda | 1 | 1 | E tarda hits |
| Staphylococcus epidermidis | 1 | 1 | S epidermidis hits |
| Synthetic construct | 1 | 1 | Synthetic construct hits |
Quantification and Characterization of OmpC T Cells Ex Vivo
To characterize the frequency and immunophenotype of OmpC-specific T cells, antigen-specific T cells were identified from peripheral blood using HLA-DRB1∗15:01 tetramers containing either OmpC or influenza hemagglutinin (Flu) peptides. Flow cytometric analysis showed that OmpC-specific T cells are found in both healthy donor and CD patients (Figure 3A, Table 3). Flu-specific T cells were used as a positive control and also were found in similar frequencies in both cohorts, although at a higher frequency than OmpC-specific cells. There was no significant difference in the frequency of peripherally circulating OmpC-specific T cells between HCs and CD patients (Figure 3B), suggesting that any difference between cohorts may be qualitative rather than quantitative. However, among CD patients for whom anti-OmpC IgA serum levels were available, a correlation was observed (Spearman rho = 0.69, P = .011) between such levels and the calculated precursor frequency of OmpC-specific T cells (Figure 3C).
Figure 3.
OmpC-specific T cells are present in both Crohn’s patients and HCs. PBMCs from CD patients (n = 24) and HCs (n = 25) were labeled with PE-conjugated tetramers, containing either OmpC 321–340 or Flu MP63 peptides, and were quantitated by flow cytometry. (A) A representative example of tetramer staining in CD and HCs. (B) Number of Flu and OmpC-specific tetramer-binding cells expressed as frequency per million CD4+ T cells. Each subject is represented by a single open circle (Flu) and closed circle (OmpC). P values are shown for Wilcoxon matched-pairs signed-rank tests of paired comparisons between Flu and OmpC-specific cells within each cohort, and for Mann–Whitney tests of unpaired comparison of Flu and OmpC-specific cells between HCs and CD patients. (C) OmpC-specific T-cell precursor frequency data from panel B is plotted against serum levels of anti-OmpC IgA for 13 CD patients from whom the latter were available. Rho and P values for nonparametric Spearman correlations are shown. ELISA, enzyme-linked immunosorbent assay.
Table 3.
Study Subject Details
| CD (n = 24) | HC (n = 39) | |
|---|---|---|
| Mean age, y (SD) | 42 (16) | 42 (14) |
| Male, % | 33 | 31 |
| Caucasian, % | 88 | 97 |
| Mean BMI (SD) | 27 (7) | 27 (5) |
| On aminosalicylate, % | 38 | 0 |
| On glucocorticoid, % | 38 | 0 |
| On immunomodulator, % | 54 | 0 |
| On anti-integrin, % | 8 | 0 |
| On anti-TNF, % | 67 | 0 |
| Bionaïve, % | 25 | 100 |
| OmpC seropositive, % | 33 | 0 |
| OmpC seronegative, % | 33 | 0 |
| OmpC unknown, % | 33 | 100 |
BMI, body mass index; TNF, tumor necrosis factor.
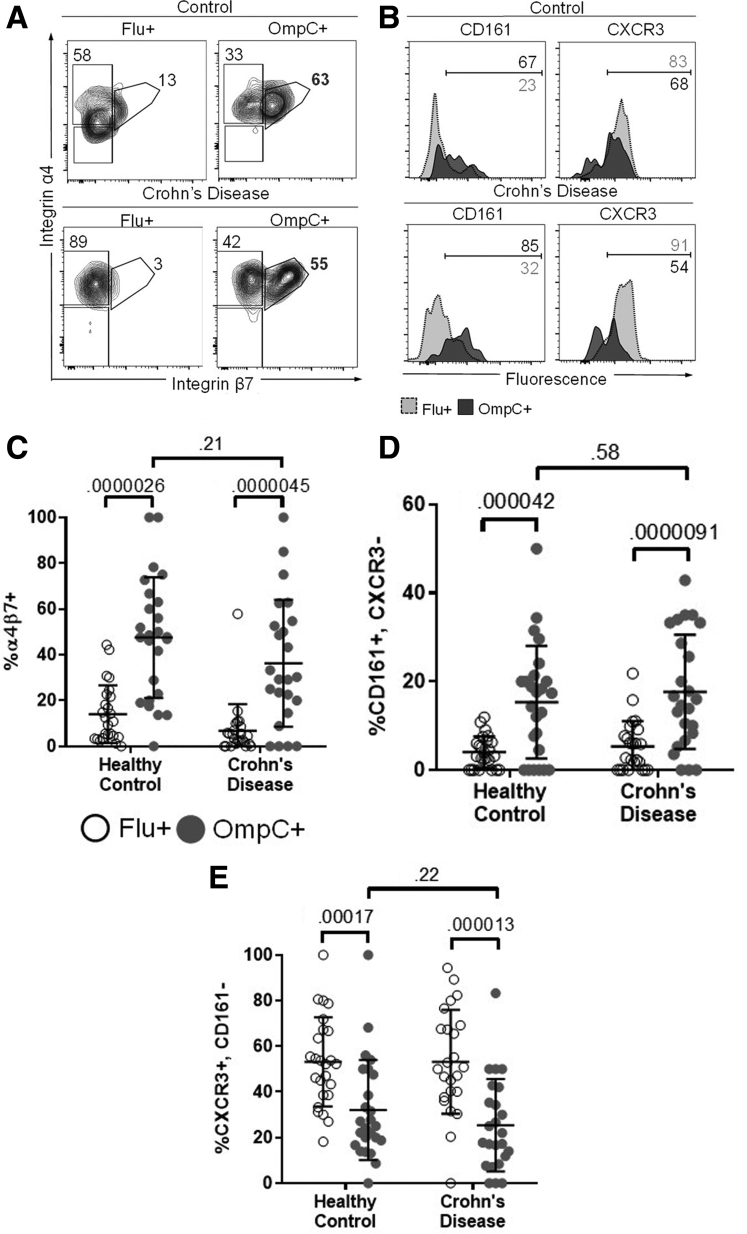
OmpC-specific T cells were found to express the gut-tropic integrin α4β7 significantly more frequently than Flu-specific T cells (mean OmpC, 43%; Flu, 10%; P = 3.8e-13, paired) (Figure 4A and C; gating strategy in Figure 5A and 5B). However, there was no difference in α4β7 expression on OmpC-specific T cells between HCs and CD patients (means: HCs, 48%; CD patients, 39%; P = .21, unpaired). OmpC-specific T cells also more frequently were CD161+/CXCR3-, a phenotype of human helper T 17 (Th17) cells,12, 13, 14, 15 than Flu-specific T cells (means: OmpC, 16.5%; Flu, 4.69%; P = 8.6e-101, paired). No significant difference in the CD161+/CXCR3- phenotype was seen between OmpC+ T cells from CD patients and HCs (means: HC, 15.3%; CD, 17.7%; P = .58, unpaired). Conversely, Flu-specific T cells more frequently were CXCR3+/CD161-, a Th1 cell phenotype,16 than OmpC-specific T cells (means: OmpC, 28.8%; Flu, 53.2%; P = 8.2e-9, paired), and again expression was not affected by disease state (Flu mean: HCs and CD patients each, 53.2%; P = .9; OmpC means: HC, 32.1%; CD, 25.4%; P = .22, unpaired). Together, these data show that regardless of disease state, peripherally circulating OmpC-specific T cells have a gut-tropic, Th17-like phenotype.
Figure 4.
Ex vivo OmpC-specific T cells have an integrin α4β7+, CD161+, CXCR3- phenotype. (A) PBMCs from CD patients (lower panels) and HCs (upper panels) were labeled with PE-conjugated OmpC (right panels) or Flu-specific tetramer (left panels), as in Figure 3, and were costained for expression of integrin α4 and β7. A representative example of α4β7 gating in Flu and OmpC-specific T cells is shown. (B) OmpC and Flu-specific T cells were co-stained with the surface markers CD161 and CXCR3. Representative histograms of CD161 and CXCR3 expression in Flu (light grey) and OmpC-specific (dark grey) T cells are shown. Percentage of Flu (white circles) and OmpC-specific (dark circles) tetramer-binding cells in 25 HC and 24 CD subjects expressing (C) the integrin α4β7 heterodimer, (D) CD161 without CXCR3, or (E) CXCR3 without CD161 is shown. P values are shown and were calculated as described in Figure 3B.
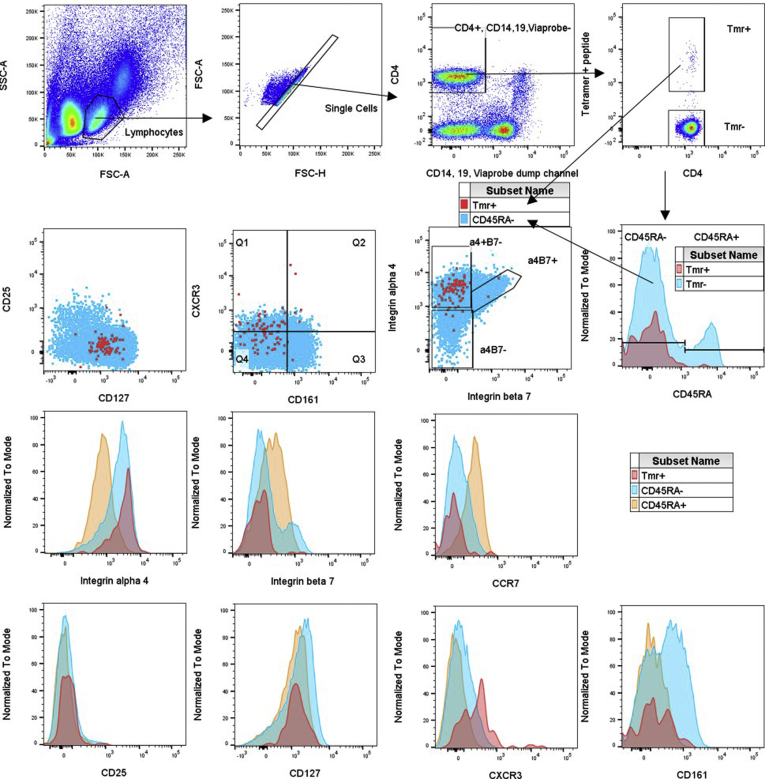
Figure 5.
Representative fluorescence-activated cell sorter (FACS) plots of ex vivo PBMCs. FACS data are shown for PBMCs from a Crohn’s patient stained with MHC II tetramers containing (A) OmpC or (B) Flu peptides. The gating strategy is denoted by arrows. Overlaid plots and histograms of CD4 T cells show tetramer-binding cells in red, CD45RA-negative cells in blue, and CD45RA+ cells in orange. FSC-A, forward scatter area; FSC-H, forward scatter height; SSC-A, side scatter area.
Clonal Expansion and Function of OmpC+ T Cells
To further assess the function of OmpC-specific T cells, we isolated, cloned, and cultured these cells from 15 HC and 15 CD patients (Figure 6). Five to 56 individual OmpC-specific T cells were sorted from each patient into independent expansion cultures, of which a small minority (∼10%) survived clonal expansion over a 2- to 3-month period.
Figure 6.
Schematic of cloning strategy. Graphic showing the steps used to generate OmpC-specific T-cell clones, as per the Materials and Methods section. The Figure was generated using BioRender software (https://biorender.com). FACS, fluorescence-activated cell sorter; PHA, phytohemagglutinin.
After clonal expansion, flow cytometry was used to exclude clones without OmpC peptide tetramer-binding, and the majority of the remaining CD4+ clones (n = 35; 15 from CD patients, 20 from HCs) were homogeneously positive for tetramer-binding (Figure 7A). Expanded OmpC-specific clones were uniformly positive for CD38, CD28, CD161, CD226, CXCR3, and Programmed cell death protein 1 (PD-1), and negative for CD45RA, CD154, and OX40, with variable expression of T cell immunoreceptor with Ig and ITIM domains (TIGIT) (Figure 8) that did not differ overall between clones from HC and CD cohorts. Differences, such as CXCR3 expression, between ex vivo and culture-expanded OmpC-specific clones presumably reflect in vitro activation.
Figure 7.
Culture-expanded OmpC-specific T cells from Crohn’s patients have less IL10 and Th2 cytokine production than those from HCs. Forty-five individual OmpC-tetramer+ T cells were sorted and successfully expanded in vitro from 8 CD patients and 9 HCs, as per the Materials and Methods section. (A) Representative fluorescence-activated cell sorter data from a clone stained with CD4 and HLA-DRB1∗15:01 tetramers loaded with OmpC 321-340 (right panel) or control peptide (left panel). (B) Each clone (unique dot) was cultured for 5 days in vitro with irradiated HLA-DRB1∗15:01+ PBMCs from a single healthy donor (unrelated to any HC from which clones were generated), with or without either OmpC 321–340 peptide or an irrelevant control peptide, and tritiated thymidine incorporation during the fifth day was measured by scintillation counting, as shown, to reflect T-cell clone proliferation. P values for the Friedman test of variation between conditions within each cohort are shown. Two CD clones that did not increase proliferation in response to OmpC peptide stimulation by more than 2 SDs above control peptide stimulation were not deemed OmpC-specific, and therefore were omitted from subsequent analyses (later). (C) Supernatants were collected from the conditions in panel B after 3 days of culture and the concentration of each of the indicated cytokines therein was measured by Luminex assay (Luminex Corporation, Austin, TX). Data are shown as a ratio of how much of each cytokine a given clone produced per unit volume in response to OmpC peptide relative to in response to control peptide. P values are shown for each cytokine for Mann–Whitney comparisons between clones from Crohn’s patients and controls. (D) Clones were cultured, as in panel B, with irradiated PBMCs presenting OmpC 321–340 peptide in the absence or presence of soluble recombinant TIGIT, 4-1BB, OX40, or CTLA-4-Fc fusion proteins, to block costimulation from CD112/CD155, 4-1BBL, OX40L, and B7-1 and -2, respectively, on antigen-presenting cells. The concentration of IL10 in the day 3 supernatants from each clone is shown for each condition, minus the basal concentration in supernatants from that clone stimulated with control peptide in the absence of costimulatory blockade. Paired comparisons of IL10 concentrations in the presence vs absence of each blockade were made by Wilcoxon signed-rank test, for which P values are shown above each column. The P value for unpaired Mann–Whitney comparison of IL10 concentrations between OmpC peptide-stimulated HCs vs CD clones in the absence of blockade is shown below the graph. (C and D) ∗P values that remained significant after Bonferroni correction for multiple comparisons. APC, antigen-presenting cell; CPM, counts per million.
Figure 8.
Representative fluorescence-activated cell sorter plots of expanded clones. Flow cytometry plots superimposing the immunophenotype of a representative in vitro–expanded, OmpC-specific, CD4 T-cell clone (red) with PBMCs from a normal healthy donor (blue). The gating strategy is denoted by arrows. FSC-A, forward scatter area; SSC-A, side scatter area.
OmpC-specific clones from healthy and CD patients were stimulated with allogenic, irradiated HLA-DRB1∗15:01+ PBMCs with or without OmpC or control peptide (from an OmpC sequence not identified as antigenic by TGEM). Clones from both HCs and CD patients proliferated in response to stimulation with irradiated antigen-presenting cell and cognate OmpC peptide. Little to no proliferation was seen with control peptide or no peptide (Figure 7B).
IL2, IL4, IL5, IL6, IL9, IL10, IL13, IL17A, IL21, IL22, tumor necrosis factor-α, and IFNγ concentrations were measured in supernatants sampled from the clonally expanded, OmpC+ cultures after 3 days of incubation. OmpC clones from CD patients produced significantly less IL10 than HC clones in response to OmpC peptide stimulation, relative to control peptide (P = 4.3e-5, unpaired) (Figure 7C). A trend toward less OmpC peptide-stimulated production of the Th2 cytokines IL4, IL5, and IL13 also was seen by clones from CD patients compared with HCs, but P values did not remain significant after correction for multiple comparisons. In contrast, clones from HCs and CD patients produced comparable amounts of IL2, IL6, IL13, IL21, tumor necrosis factor-α, and IFN-γ, showing that the earlier-described defect in IL10 production by CD clones does not reflect a global defect in their antigen-responsive cytokine production. Little to no IL17A or IL22 was detected from most clones in response to OmpC. Although a minority (23%) of HC clones made more than 10-fold baseline levels of IL9 in response to OmpC peptide, this cytokine also was minimally and heterogeneously produced by most clones.
The absolute concentration of IL10 in supernatants from OmpC peptide-stimulated clones significantly increased in the presence of a soluble recombinant TIGIT-fragment crystallizable region (Fc) molecule for clones from HCs (P = 1.6e-3, paired) but not CD patients (P = .98, paired) (Figure 7D). TIGIT-Fc presumably blocks TIGIT/CD226 co-stimulation via CD155 and/or CD112 molecules on OmpC peptide-presenting cells. Blockade of 4-1BB ligand (4-1BBL, CD137L) with soluble recombinant 4-1BB-Fc significantly increased IL10 production by both CD and HC clones. Blockade of B7-1/B7-2 costimulation (with soluble recombinant cytotoxic T-lymphocyte-associated protein 4 [CTLA4-Fc] or OX40 ligand (OX40L, CD134L) (with soluble recombinant OX40-Fc) had little effect on IL10 secretion.
T-Cell Receptor Sequences of OmpC+ T-Cell Clones
To evaluate the clonality of OmpC-specific T cells, complementary DNA (cDNA) was generated from each of 35 clones, expanded as described earlier, and the T-cell receptor (TCR) α and β chains were sequenced. Despite being HLA-DRB1∗15:01-restricted T cells recognizing the same peptide antigen, there was considerable heterogeneity in TCR variable domain use among clones (Figure 9A). The predominant V-region alleles were TRAV19∗01 for α chains and TRBV24-1 for β chains, with both being expressed together in 6 of the clones (2 from a CD patient, 4 from healthy controls). However, similar or identical amino acid sequences were observed frequently in the hypervariable third complementarity determining regions (CDR3) of these genes (Figure 9B), where TCR–peptide interactions occur. In at least 1 case, an identical α and β chain pairing was seen in 2 different T-cell clones (19B7 and 19D7) isolated from the same donor (donor 19, a healthy donor). This suggests that a high enough clonal precursor frequency, and hence clonal expansion, in the peripheral blood of this individual for 2 sister T cells was found among the few OmpC-specific T cells (17, in the case of donor 19) that were sorted from this individual. However, in most cases different clones isolated from any given donor showed differences in their α and/or β CDR3 sequence, to indicate that OmpC-specific T cells are polyclonal (or at least oligoclonal).
Figure 9.
OmpC-specific T-cell clones show a high frequency of public TCR α and β chains. Each of 34 expanded OmpC-specific clone had their TCR α and β chain genes sequenced in their entirety. (A) The number of clones from CD (n = 8) and HC (n = 9) donors with each of the indicated α and β TCR V-regions is shown. (B) For the α and β TCR gene of each clone, the V, J, and (in the case of β chains) D regions used is shown, as well as the CDR3 amino acid sequence unique to that clone. In the latter, X represents an amino acid whose character was unclear from nucleotide sequence data, and hyphens have been inserted to best align amino acid motifs with one another, using SALIGN (https://modbase.compbio.ucsf.edu/salign). Beneath these are graphic depictions of aligned CDR3 consensus motifs (made with https://weblogo.berkeley.edu/logo.cgi). Rows for clones with identical α and/or β chain sequences have been placed adjacent to one another and identical α and/or β chain sequences have been highlighted with the same color. NA, unsuccessful sequencing reactions.
Curiously, despite having sequenced the TCRs of fewer than 3 dozen T-cell clones, we observed multiple examples of the identical amino acid sequence appearing in the α and/or β chain of clones obtained from different people, or even different cohorts (ie, healthy vs Crohn’s). The exact same amino acid sequence for both the α and β chains was identified in clones 22C8 (from Crohn’s subject 22), 9B8 and 10B3 (from subjects 9 and 10, respectively, both HCs). Thus, although the OmpC-responsive T cells we identified represent a polyclonal population, frequent public TCR sequences recur in different HLA-DRB1∗15:01+ individuals, regardless of whether or not they have CD.
Discussion
This report documents the identification of a tolerogenic defect in a specific peptide T-cell epitope from an intestinal commensal antigen (OmpC) against which some Crohn’s patients are known to show aberrant humoral immunity. This discovery enabled a reductionist approach to evaluating T-cell tolerance to intestinal flora by eliminating the need for in vitro T-cell activation to detect antigen-specific cells. By using MHC II tetramers, we were able to examine T cells ex vivo, without introducing intact antigen or requiring in vitro processing by antigen-presenting cells. This approach showed OmpC-specific cells to express the gut-homing integrin α4β7 disproportionately,17 as one might predict for cells recognizing intestinal flora. In addition, we found OmpC-specific cells express CD161, a marker for Th17 cells,12, 13, 14, 15 which also is expressed more commonly by intestinal than peripheral CD4+ T cells.18 Because Th17 cells are associated with neutrophilic immune responses to bacterial pathogens,19 the expression of a Th17 marker by T cells specific for an E coli antigen also might be expected. This phenotype significantly contrasted with CXCR3 expression, and thus Th1-like phenotype, we found in peripheral T cells specific for a known peptide antigen of influenza from the same subjects. Our OmpC 321-340 peptide sequence aligned 100% when examined via NCBI protein BLAST with Enterobacteraceae including E coli, Shigella, Klebsiella, and Salmonella, which has been shown repeatedly to have increased abundance in IBD patients.20,21
Among the markers we analyzed, we did not identify significant differences in the ex vivo phenotype or precursor frequency of OmpC-specific T cells between CD patients and HCs. However, we did find a correlation between the precursor frequency of OmpC-specific T cells and the serum levels of anti-OmpC IgA in the CD patients for whom the latter was available, supporting our initial hypothesis that these T cells provide help for B cells making antibodies against, and hence breaking serologic immune tolerance to, normal commensal flora. Because our analyses were restricted to peripheral blood, it is possible that sequestration of OmpC-specific T cells to the intestinal mucosa or mesenteric lymph nodes could have obscured quantitative differences between Crohn’s patients and controls. Indeed, the intestinal mucosa and lymphatics are presumably where OmpC-specific T and B cells interact with E coli OmpC antigen. Unfortunately, our efforts to use MHC II tetramers with mucosal lymphocytes from the colonic lamina propria were technically unsuccessful, perhaps as a consequence of the extended preparation and collagenase treatment necessary to extract lymphocytes from intact tissue.
Although the ex vivo phenotype of OmpC-specific T cells showed few specific differences between HCs and CD patients, in vitro–expanded OmpC-specific T-cell clones showed a striking difference between the 2 cohorts, with clones from HCs showing significantly more antigen-responsive IL10 production than those from CD patients. IL10 has long been known to be a critical cytokine for immunoregulation, particularly in the gastrointestinal tract, because mice lacking the IL10 gene develop a severe chronic enterocolitis.22 Similarly, human beings born with genetic defects in the IL10 receptor (CDW210a) develop a very early and severe form of Crohn’s disease,23 and genetic polymorphisms in the IL10 gene have been linked to Crohn’s disease.24 However, most CD patients do not show a global deficiency in IL10 production,25,26 and therapies based on IL10 replacement in CD have been unsuccessful.27 Our data show that, in CD, there is defective IL10 production by gut-tropic (eg, integrin α4β7+) peripheral T cells specifically recognizing a gut flora antigen peptide. If this defect is limited to T cells specific for normal gut commensals, it would have been obscured in prior analyses that could not take antigen specificity into account.
IL10 production by OmpC peptide-stimulated clones from HCs paradoxically was increased by blockade of the ligands for the T-cell costimulatory receptors 4-1BB (CD137) and CD226 (by soluble Fc fusion proteins of 4-1BB and TIGIT, respectively), suggesting that these costimulatory signals normally suppress IL10 production by commensal-specific T cells. However, Fc-TIGIT failed to further boost IL10 production in OmpC-specific T-cell clones from CD patients, suggesting that any inhibitory signal from CD226 on IL10 production is either constitutively active or redundant in CD. Thus, in the absence of CD226 ligation, the difference in IL10 production between OmpC-specific T cells from HCs vs CD patients was increased further. Because TIGIT may act as an inhibitory receptor on T cells by competing with CD226 for its ligands (CD155 and CD112), our data suggest that commensal-specific T cells in CD may be abnormally refractory to the regulatory activity of TIGIT. This may be particularly important in the bowel, where epithelial cells express copious CD15528 as a ligand for both TIGIT and CD226.
In addition to IL10, OmpC-specific T-cell clones from CD patients showed reduced production of the Th2 cytokines IL4, IL5, and IL13. Although none of these differences was significant enough to withstand correction for multiple comparisons, they collectively support the consensus that there is a shift in the immune system away from Th2 differentiation in favor of Th1 and Th17 differentiation in CD.
Despite expressing the Th17 marker CD161 ex vivo, OmpC-specific T-cell clones were found to make little or no IL17A, IL21, or IL22 after in vitro expansion. This is in contrast to findings described by Hegazy et al29 in which CD154-based bacterial-specific T cells were isolated from peripheral blood of healthy and IBD patients and found to have robust IL17A production despite disease status. These differences in findings could reflect a selection against Th17 cells in culture because only a minority of sorted single cells survived the clonal expansion process. To avoid biasing cell phenotypes in vitro, we did not add exogenous IL23, which is known to support Th17 survival, and may not have been produced in sufficient amounts by feeder cells in expansion cultures to maintain Th17 cells. Similarly, Hegazy et al29 found the addition of IL1β, IL6, or IL23 during bacterial-specific T-cell stimulation led to a 1.5- to 2-fold increase in IL17A production. Alternatively, Th17 cells may have changed their phenotype in culture. Indeed, bacteria-specific Th17 cells have been shown to transiently lose IL17A and gain IL10 production after in vitro culture, via an IL2-dependent mechanism that could be reversed with IL1β.30 Because copious exogenous IL2 was added to our cultures throughout expansion, it is possible that a loss of IL17A production was stabilized in our OmpC-specific clones, although this does not explain why the accompanying expression of IL10 was seldom seen in clones from Crohn’s patients.
Although only a relatively small number (35) of expanded OmpC peptide-specific T-cell clones had their TCR α and/or β chain successfully sequenced, a surprising fraction of the amino acid sequences (11% of α sequences, 19% of β sequences) were found in more than 1 person, regardless of whether or not subjects had CD. In human beings, such public TCR sequences, shared between unrelated individuals, can be found among CD8+ T cells,31 and are enriched within the antigen-specific repertoires thereof in Epstein–Barr virus,32 cytomegalovirus,33 and influenza A infection.34 Although far less is known about public TCRs in CD4+ T cells, rare individuals capable of spontaneously controlling human immunodeficiency virus infection have been reported to harbor a high frequency of public clonotypes among their human immunodeficiency virus Gag295-specific CD4+ T cells,35 which can recognize their cognate antigenic peptide in a variety of different HLA molecules.36 In addition, a recent study of patients with celiac disease, another immune-mediated disease of the gastrointestinal tract, reported that 10% of the gluten-specific T-cell repertoire comprised public TCRs.37
Future studies may determine if the OmpC-specific clones we generated can recognize their cognate peptide antigen in MHC molecules other than HLA-DRB1∗15:01, or if their public TCR α or β chain sequences can be found among the T cells of individuals without HLA-DRB1∗15:01. However, none of the public TCR sequences we identified in OmpC-specific clones were among a published list of more than 10,000 public β chain amino acid sequences shared by at least 2 individuals in a reference cohort of 39 healthy Russian Caucasians.38 This suggests that these OmpC clones are indeed HLA-restricted, are determined by regional/geographic factors, and/or are simply not as common as other public clonotypes, which may be dominated by CD8+ cells.
In summary, this report details a tetramer-based reductionist approach to identifying circulating CD4+ T cells specific for a known antigen (OmpC) of a common intestinal commensal bacteria (E coli). In HLA-DRB1∗15:01+ CD patients, the frequency of such OmpC-specific T cells correlated with titers of the IgA antibodies against OmpC that commonly are increased in CD patients. Relative to healthy subjects, we found no quantitative difference in the frequency of these T cells in people with CD, but rather a qualitative defect in their ability to produce IL10 after in vitro expansion from CD donors. This suggests that an antigen-specific immunoregulatory defect is present in CD, which may be fundamental to its immunopathogenesis. We also found a surprisingly high frequency of public TCR sequences among the few dozen OmpC-specific T-cell clones we successfully expanded and sequenced, suggesting that the repertoire of responses to the commensal E coli is limited and conserved between HLA-DRB1∗15:01+ individuals in a manner previously observed with viral infections. Future studies may help to determine how such conserved responses are generated and play a role in regulating the immune response to normal intestinal flora.
Methods
All authors had access to the study data and reviewed and approved the final manuscript.
Study Subject Genotyping, Serotyping, and Selection
DNA from 36 patients with CD (age, 20–66 y; 59% female) in the Benaroya Research Institute IBD Biorepository with known OmpC or CBir1 seropositivity from medical records underwent HLA DRB1 genotyping using sequence-specific oligonucleotide primers with low-resolution Unitray SSP Kits (Invitrogen, Carlsbad, CA), reflected in Figure 1 and Table 1. DNA from a subsequent cohort of CD patients then was genotyped specifically for HLA-DRB1∗15:01 using the high-resolution, sequence-specific oligonucleotide primers with Unitray SSP Kits (Invitrogen). Twenty-four HLA-DRB1∗15:01+ CD patients were selected for all subsequent studies, for which an age- and sex-matched cohort of HLA-DRB1∗15:01+ HCs (previously genotyped) was identified from the control Biorepository at our institute. Serum from some of these CD patients was evaluated previously for the presence of IgA antibodies to OmpC via enzyme-linked immunosorbent assay as previously described1 (courtesy of Dr Dermott McGovern, Cedars-Sinai Medical Center, Los Angeles, CA). Demographic and clinical data are summarized in Table 3. This study protocol was approved by the Ethics Committee at Benaroya Research Institute and was performed in accordance with the principles stated in the Declaration of Helsinki. All patients signed an informed consent form before inclusion in the study.
PBMC Isolation
PBMCs were isolated from fresh blood using Lymphoprep (Axis-Shield, Oslo, Norway) density gradient. When necessary, PBMCs were frozen in liquid nitrogen in 7% dimethyl sulfoxide in calf serum and stored in liquid nitrogen (-120°C) until time of use.
TGEM
TGEM was conducted as previously described11 to identify HLA-DRB1∗15:01-restricted peptide epitopes within the mature amino acid sequence (lacking the predicted 16 amino acid leader sequence) encoded by the E coli OmpC gene (uniprot.org, entry P06996). Briefly, we obtained a peptide library of 44 peptides of 20 amino acids each, overlapping by 12 amino acids, which spanned the entire 367 amino acids encoded by the E coli OmpC gene. Four million fresh PBMCs per well (9 wells total) from HLA-DRB1∗15:01+ donors were stimulated in 48-well plates with pools of 4–5 consecutive and overlapping peptides. After 14 days in culture, supplemented with IL2 on days 7 and 10, approximately 2 × 105 cells in 100 uL were stained for 60 minutes at 37°C with phycoerythrin (PE)-conjugated HLA-DRB1∗15:01 tetramers previously loaded with a mix of the peptides of their respective pools. Subsequently, cells were stained with CD4-APC (clone RPA-T4; BD Biosciences, Mountainview, CA), CD3-peridinin-chlorophyll-protein complex (PerCP) (clone SP34-2; BioLegend, San Diego, CA), and CD25–fluorescein isothiocyanate (FITC) monoclonal antibodies (clone BC96; eBioscience, San Diego, CA) and analyzed by flow cytometry. Cultures that showed positive tetramer staining with their respective peptide pool subsequently were divided into 4 or 5 aliquots, each of which was stained and analyzed with tetramers loaded with the individual peptides of that pool to show the single peptide(s) recognized within the pool. Tetramers were assembled by conjugating biotinylated HLA-DRB1∗15:01 molecules with PE-conjugated streptavidin and loading with peptides. MP63, an influenza (flu) epitope, was used as a control tetramer because it is a validated antigen with high levels of response in the community.39
Flow Cytometry
Frozen PBMCs from CD patients and HC donors were thawed and incubated at 37°C with 1 μmol/L dasatinib (Bristol-Myers Squibb, New York, NY) for 8 minutes to prevent TCR down-regulation. Samples then were incubated with OmpC–PE–labeled tetramer for 90 minutes at room temperature. Tetramer enrichment was performed using the Miltenyi (Cologne, Germany) OctoMACS separation system with anti-PE beads. A total of 50 μL from the pre-enrichment samples was obtained to calculate tetramer frequency (described later). Samples were stained with appropriate monoclonal antibody mixtures (integrin β7-FITC [clone FIB504; eBioscience, San Diego, CA], CD127-BV650 [clone A019D5; eBioscience], CD4-APC-e780 [clone RPA-T4; eBioscience], Viaprobe-PerCP-Cy5.5 [Via-Probe Cell Viability Solution; BD Biosciences, Franklin Lakes, NJ], CD49d-BV510 [clone 9F10; BD Biosciences], CD45RA-BV605 [clone HI100; BD Biosciences], CCR7-A700 [clone 150503; BD Biosciences], CXCR3-PE-Cy7 [clone 1C6/CXCR3; BD Biosciences], CD14-PerCP-Cy5.5 [clone HCD14; BioLegend, San Diego, CA], CD161-BV421 [clone HP-3G10; BioLegend], CD19-PerCP-Cy5.5 [clone HIBI9; BioLegend], CD25-APC [clone BC96; BioLegend], and tetramer-OmpC-PE). Live CD4+, CD45RA-, OmpC-specific T cells were identified and characterized on an Aria II flow cytometer (BD Biosciences) and analyzed using FlowJo software (FlowJo LLC, Ashland, OR).
Single-Cell Sorting, Cloning, and Proliferation Assays
Single-cell sorting was performed after tetramer enrichment, as described earlier, and staining with the following antibody cocktail: CD226-AF647, CCR7-AF700, CD4-APC, CD14-FITC, CD19-FITC, live/dead green, TIGIT-PerCP-eF710, CCR10-BV421, CD45RA-BV510, CD103-BV605, CD28-BV650, OmpC-PE, CRTh2-PE-dazzle594, and CCR4-PE-Cy7. Single, live CD4+CD45RA- OmpC-specific T cells were sorted into separate wells of a 96-well, round-bottom plate, each of which contained 175 μL of 30% complete human sera RPMI and fresh, irradiated (5000 rad) HLA-mismatched healthy donor PBMC feeder cells to facilitate growth. Cultures were incubated at 37°C, and IL2 (2 U/well) plus phytohemagglutinin (5 μg/mL) were added on day 2 for stimulation. On day 14, media was exchanged and again freshly obtained PBMCs from HLA-mismatched healthy donors were irradiated and fed to cells with IL2 and phytohemagglutinin. Day 28 media exchange was performed with fresh HLA-matched (DRB1501+) PBMC feeder cells that were pulsed with OmpC peptide (10 μg/mL for 1 hour at 37°C) and then irradiated before being added to single-cell culture. This alternating pattern continued for a total of 2 cycles until adequate clonal proliferation was seen. Low-dose (5 μg/mL) Amphotericin B was added intermediately to cultures to prevent fungal contamination (Figure 6).
After 8–12 weeks of clonal expansion, OmpC specificity was confirmed by flow cytometry using OmpC peptide-loaded tetramers, as described earlier. Seventeen cultures showing less than 24% tetramer binding were discarded. Thirty-five of the remaining 45 clones showed at least 75% tetramer binding, with more than half being more than 90% OmpC+. Functional specificity was confirmed by culturing cloned T cells with irradiated allogeneic HLA-DRB1∗15:01+ PBMCs loaded with OmpC peptide, control peptide, or nothing, in the presence or absence of soluble recombinant CTLA4-Fc, OX40-Fc, 4-1BB-Fc, or TIGIT-Fc to block their costimulatory receptors on antigen-presenting cells. Supernatants were harvested from cultures after 1 or 3 days of culture, frozen, and later analyzed for cytokine content by multiplex assay (eBioscience). After 3 days of culture, tritiated thymidine was added to cultures, and cells were harvested on the fourth day of culture to measure thymidine incorporation by scintillation counting.
TCR Sequencing
Messenger RNA from in vitro–expanded OmpC-specific T-cell clones, described earlier, was amplified and sequenced according to 1 of 2 protocols.
RNA from 500,000 clonal cells was isolated using QIAzol Lysis Reagent (Qiagen, Hilden, Germany) according to the manufacturer’s guidelines. A total of 0.4–1.0 μg of RNA was converted to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). The cDNA product was diluted 10-fold with nuclease-free water and 1 μL was used for multiplex and single polymerase chain reaction experiments. Primer sets for the human T-cell receptor α and β chains spanning the variable to constant regions were synthesized by Invitrogen (Carlsbad, CA) according to published sequences.40 Primer pools were organized for multiplex reactions and assessed using a Multiple Primer Analyzer tool (https://www.thermofisher.com/us/en/home/brands/thermo-scientific/molecular-biology/molecular-biology-learning-center/molecular-biology-resource-library/thermo-scientific-web-tools/multiple-primer-analyzer.html) from Thermo-Fisher Scientific (Waltham, MA) to avoid cross primer dimerization. cDNA was amplified with primer pools using the Phusion High-Fidelity DNA Polymerase Kit (Thermo Fisher Scientific) and viewed by gel electrophoresis. After screening cDNA in the primer pools, cDNA then was amplified using the individual primer pairs from pools that contained product. Positive products were purified using the QIAquick Gel Extraction Kit (Qiagen) and sent to Eurofins (Luxembourg City, Luxembourg) Genomics for Sanger sequencing using the known forward variable region primer.
To use fewer cells and obviate RNA isolation, an additional protocol was devised where approximately 100–200 clonal cells were suspended in nuclease-free water and immediately added to a cDNA reaction. The TCR α and β messenger RNA then was amplified and sequenced according to described methods.41 To check quality control, a subset of clone TCRs were sequenced by both of the earlier-described methods and generated identical sequences.
Sequences were aligned to reference directories using the IMGT/V-QUEST tool to determine the V(D)J recombination (http://www.imgt.org/IMGT_vquest/vquest).42,43
Statistical Analyses
Frequency of tetramer+ cells was calculated by dividing the number of tetramer+ CD4+ cells after tetramer enrichment by the number of total live CD4 T cells analyzed before enrichment multiplied by the dilution factor.
Data were not assumed to have a Gaussian distribution, so 2-tailed Mann–Whitney U tests were performed for unpaired 2-group comparisons, Wilcoxon signed-rank tests were performed for paired 2-way analyses, and nonparametric Spearman correlation was performed for linked continuous data sets. Three-way paired comparison in Figure 5B was performed with a Friedman test. All statistical analyses were performed using Excel (Microsoft, Redmond, WA) spreadsheets and Prism (GraphPad, San Diego, CA). Error bars show the medians and interquartile ranges. For analyses involving multiple comparisons, a Bonferroni correction was performed to determine a P value threshold for significance.
Acknowledgments
The authors wish to thank Kassidy Benoscek for patient recruitment, Thien-Son Nguyen for specimen curation, and “Aru” K. Arumuganathan for assistance with flow cytometry (Benaroya Research Institute). The authors also would like to thank Dermott McGovern at Cedars Sinai Medical Center, Los Angeles, CA, for measuring anti-OmpC IgA titers in a subset of CD patients.
All reagents used in this manuscript are commercially available to the public through the indicated vendors. All data associated with this study are available in the main text or the Supplementary Materials.
CRediT Authorship Contributions
Amiko M. Uchida, MD (Data curation: Equal; Investigation: Lead; Writing – original draft: Lead); Elisa K. Boden, MD (Data curation: Equal; Formal analysis: Supporting; Investigation: Supporting; Project administration: Supporting; Supervision: Supporting; Writing – review & editing: Supporting); Eddie A. James, PhD (Conceptualization: Supporting; Investigation: Supporting; Methodology: Supporting; Resources: Supporting; Supervision: Supporting; Writing – review & editing: Supporting); Donna M. Shows (Data curation: Supporting; Investigation: Supporting; Methodology: Lead); Andrew J. Konecny (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting); James Daniel Lord, MD, PhD (Conceptualization: Lead; Data curation: Equal; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Supporting; Project administration: Lead; Resources: Lead; Supervision: Lead; Visualization: Lead; Writing – review & editing: Lead).
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was made possible by pilot grants from The Rainin Foundation and The Digestive Disease Institute at Virginia Mason.
References
- 1.Landers C.J., Cohavy O., Misra R., Yang H., Lin Y.C., Braun J., Targan S.R. Selected loss of tolerance evidenced by Crohn's disease-associated immune responses to auto- and microbial antigens. Gastroenterology. 2002;123:689–699. doi: 10.1053/gast.2002.35379. [DOI] [PubMed] [Google Scholar]
- 2.Targan S.R., Landers C.J., Yang H., Lodes M.J., Cong Y., Papadakis K.A., Vasiliauskas E., Elson C.O., Hershberg R.M. Antibodies to CBir1 flagellin define a unique response that is associated independently with complicated Crohn's disease. Gastroenterology. 2005;128:2020–2028. doi: 10.1053/j.gastro.2005.03.046. [DOI] [PubMed] [Google Scholar]
- 3.Kaur M., Panikkath D., Yan X., Liu Z., Berel D., Li D., Vasiliauskas E.A., Ippoliti A., Dubinsky M., Shih D.Q., Melmed G.Y., Haritunians T., Fleshner P., Targan S.R., McGovern D.P. Perianal Crohn's disease is associated with distal colonic disease, stricturing disease behavior, IBD-associated serologies and genetic variation in the JAK-STAT pathway. Inflamm Bowel Dis. 2016;22:862–869. doi: 10.1097/MIB.0000000000000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrante M., Henckaerts L., Joossens M., Pierik M., Joossens S., Dotan N., Norman G.L., Altstock R.T., Van Steen K., Rutgeerts P., Van Assche G., Vermeire S. New serological markers in inflammatory bowel disease are associated with complicated disease behaviour. Gut. 2007;56:1394–1403. doi: 10.1136/gut.2006.108043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papp M., Altorjay I., Dotan N., Palatka K., Foldi I., Tumpek J., Sipka S., Udvardy M., Dinya T., Lakatos L., Kovacs A., Molnar T., Tulassay Z., Miheller P., Norman G.L., Szamosi T., Papp J., Hungarian I.B.D.S.G., Lakatos P.L. New serological markers for inflammatory bowel disease are associated with earlier age at onset, complicated disease behavior, risk for surgery, and NOD2/CARD15 genotype in a Hungarian IBD cohort. Am J Gastroenterol. 2008;103:665–681. doi: 10.1111/j.1572-0241.2007.01652.x. [DOI] [PubMed] [Google Scholar]
- 6.Ma C.S., Deenick E.K., Batten M., Tangye S.G. The origins, function, and regulation of T follicular helper cells. J Exp Med. 2012;209:1241–1253. doi: 10.1084/jem.20120994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanders C.J., Yu Y., Moore D.A., 3rd, Williams I.R., Gewirtz A.T. Humoral immune response to flagellin requires T cells and activation of innate immunity. J Immunol. 2006;177:2810–2818. doi: 10.4049/jimmunol.177.5.2810. [DOI] [PubMed] [Google Scholar]
- 8.Shen C., Landers C.J., Derkowski C., Elson C.O., Targan S.R. Enhanced CBir1-specific innate and adaptive immune responses in Crohn's disease. Inflamm Bowel Dis. 2008;14:1641–1651. doi: 10.1002/ibd.20645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calderon-Gomez E., Bassolas-Molina H., Mora-Buch R., Dotti I., Planell N., Esteller M., Gallego M., Marti M., Garcia-Martin C., Martinez-Torro C., Ordas I., Singh S., Panes J., Benitez-Ribas D., Salas A. Commensal-specific CD4(+) cells from patients with Crohn's disease have a T-helper 17 inflammatory profile. Gastroenterology. 2016;151:489–500 e3. doi: 10.1053/j.gastro.2016.05.050. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi F., Smith K.D., Ozinsky A., Hawn T.R., Yi E.C., Goodlett D.R., Eng J.K., Akira S., Underhill D.M., Aderem A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 11.Archila L.L., Kwok W.W. Tetramer-guided epitope mapping: a rapid approach to identify HLA-restricted T-cell epitopes from composite allergens. Methods Mol Biol. 2017;1592:199–209. doi: 10.1007/978-1-4939-6925-8_16. [DOI] [PubMed] [Google Scholar]
- 12.Cosmi L., De P.R., Santarlasci V., Maggi L., Capone M., Frosali F., Rodolico G., Querci V., Abbate G., Angeli R., Berrino L., Fambrini M., Caproni M., Tonelli F., Lazzeri E., Parronchi P., Liotta F., Maggi E., Romagnani S., Annunziato F. Human interleukin 17-producing cells originate from a CD161+CD4+ T cell precursor. J Exp Med. 2008;205:1903–1916. doi: 10.1084/jem.20080397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Annunziato F., Cosmi L., Santarlasci V., Maggi L., Liotta F., Mazzinghi B., Parente E., Fili L., Ferri S., Frosali F., Giudici F., Romagnani P., Parronchi P., Tonelli F., Maggi E., Romagnani S. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204:1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleinschek M.A., Boniface K., Sadekova S., Grein J., Murphy E.E., Turner S.P., Raskin L., Desai B., Faubion W.A., de Waal M.R., Pierce R.H., McClanahan T., Kastelein R.A. Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J Exp Med. 2009;206:525–534. doi: 10.1084/jem.20081712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maggi L., Santarlasci V., Capone M., Peired A., Frosali F., Crome S.Q., Querci V., Fambrini M., Liotta F., Levings M.K., Maggi E., Cosmi L., Romagnani S., Annunziato F. CD161 is a marker of all human IL-17-producing T-cell subsets and is induced by RORC. Eur J Immunol. 2010;40:2174–2181. doi: 10.1002/eji.200940257. [DOI] [PubMed] [Google Scholar]
- 16.Bonecchi R., Bianchi G., Bordignon P.P., D'Ambrosio D., Lang R., Borsatti A., Sozzani S., Allavena P., Gray P.A., Mantovani A., Sinigaglia F. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J Exp Med. 1998;187:129–134. doi: 10.1084/jem.187.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker C.M., Cepek K.L., Russell G.J., Shaw S.K., Posnett D.N., Schwarting R., Brenner M.B. A family of beta 7 integrins on human mucosal lymphocytes. Proc Natl Acad Sci U S A. 1992;89:1924–1928. doi: 10.1073/pnas.89.5.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lord J., Chen J., Thirlby R.C., Sherwood A.M., Carlson C.S. T-cell receptor sequencing reveals the clonal diversity and overlap of colonic effector and FOXP3+ T cells in ulcerative colitis. Inflamm Bowel Dis. 2015;21:19–30. doi: 10.1097/MIB.0000000000000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stockinger B., Veldhoen M. Differentiation and function of Th17 T cells. Curr Opin Immunol. 2007;19:281–286. doi: 10.1016/j.coi.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 20.Baumgart M., Dogan B., Rishniw M., Weitzman G., Bosworth B., Yantiss R., Orsi R.H., Wiedmann M., McDonough P., Kim S.G., Berg D., Schukken Y., Scherl E., Simpson K.W. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 21.Butto L.F., Schaubeck M., Haller D. Mechanisms of microbe-host interaction in Crohn's disease: dysbiosis vs. pathobiont selection. Front Immunol. 2015;6:555. doi: 10.3389/fimmu.2015.00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn R., Lohler J., Rennick D., Rajewsky K., Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 23.Glocker E.O., Kotlarz D., Boztug K., Gertz E.M., Schaffer A.A., Noyan F., Perro M., Diestelhorst J., Allroth A., Murugan D., Hatscher N., Pfeifer D., Sykora K.W., Sauer M., Kreipe H., Lacher M., Nustede R., Woellner C., Baumann U., Salzer U., Koletzko S., Shah N., Segal A.W., Sauerbrey A., Buderus S., Snapper S.B., Grimbacher B., Klein C. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franke A., McGovern D.P., Barrett J.C., Wang K., Radford-Smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R., Anderson C.A., Bis J.C., Bumpstead S., Ellinghaus D., Festen E.M., Georges M., Green T., Haritunians T., Jostins L., Latiano A., Mathew C.G., Montgomery G.W., Prescott N.J., Raychaudhuri S., Rotter J.I., Schumm P., Sharma Y., Simms L.A., Taylor K.D., Whiteman D., Wijmenga C., Baldassano R.N., Barclay M., Bayless T.M., Brand S., Buning C., Cohen A., Colombel J.F., Cottone M., Stronati L., Denson T., de V.M., D'Inca R., Dubinsky M., Edwards C., Florin T., Franchimont D., Gearry R., Glas J., Van G.A., Guthery S.L., Halfvarson J., Verspaget H.W., Hugot J.P., Karban A., Laukens D., Lawrance I., Lemann M., Levine A., Libioulle C., Louis E., Mowat C., Newman W., Panes J., Phillips A., Proctor D.D., Regueiro M., Russell R., Rutgeerts P., Sanderson J., Sans M., Seibold F., Steinhart A.H., Stokkers P.C., Torkvist L., Kullak-Ublick G., Wilson D., Walters T., Targan S.R., Brant S.R., Rioux J.D., D'Amato M., Weersma R.K., Kugathasan S., Griffiths A.M., Mansfield J.C., Vermeire S., Duerr R.H., Silverberg M.S., Satsangi J., Schreiber S., Cho J.H., Annese V., Hakonarson H., Daly M.J., Parkes M. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kucharzik T., Stoll R., Lugering N., Domschke W. Circulating antiinflammatory cytokine IL-10 in patients with inflammatory bowel disease (IBD) Clin Exp Immunol. 1995;100:452–456. doi: 10.1111/j.1365-2249.1995.tb03721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akagi S., Hiyama E., Imamura Y., Takesue Y., Matsuura Y., Yokoyama T. Interleukin-10 expression in intestine of Crohn disease. Int J Mol Med. 2000;5:389–395. doi: 10.3892/ijmm.5.4.389. [DOI] [PubMed] [Google Scholar]
- 27.Marlow G.J., van Gent D., Ferguson L.R. Why interleukin-10 supplementation does not work in Crohn's disease patients. World J Gastroenterol. 2013;19:3931–3941. doi: 10.3748/wjg.v19.i25.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwasaki A., Welker R., Mueller S., Linehan M., Nomoto A., Wimmer E. Immunofluorescence analysis of poliovirus receptor expression in Peyer's patches of humans, primates, and CD155 transgenic mice: implications for poliovirus infection. J Infect Dis. 2002;186:585–592. doi: 10.1086/342682. [DOI] [PubMed] [Google Scholar]
- 29.Hegazy A.N., West N.R., Stubbington M.J.T., Wendt E., Suijker K.I.M., Datsi A., This S., Danne C., Campion S., Duncan S.H., Owens B.M.J., Uhlig H.H., McMichael A., Oxford I.B.D.C.I., Bergthaler A., Teichmann S.A., Keshav S., Powrie F. Circulating and tissue-resident CD4(+) T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology. 2017;153:1320–1337 e16. doi: 10.1053/j.gastro.2017.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zielinski C.E., Mele F., Aschenbrenner D., Jarrossay D., Ronchi F., Gattorno M., Monticelli S., Lanzavecchia A., Sallusto F. Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature. 2012;484:514–518. doi: 10.1038/nature10957. [DOI] [PubMed] [Google Scholar]
- 31.Robins H.S., Srivastava S.K., Campregher P.V., Turtle C.J., Andriesen J., Riddell S.R., Carlson C.S., Warren E.H. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010;2:47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Argaet V.P., Schmidt C.W., Burrows S.R., Silins S.L., Kurilla M.G., Doolan D.L., Suhrbier A., Moss D.J., Kieff E., Sculley T.B., Misko I.S. Dominant selection of an invariant T cell antigen receptor in response to persistent infection by Epstein-Barr virus. J Exp Med. 1994;180:2335–2340. doi: 10.1084/jem.180.6.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trautmann L., Rimbert M., Echasserieau K., Saulquin X., Neveu B., Dechanet J., Cerundolo V., Bonneville M. Selection of T cell clones expressing high-affinity public TCRs within human cytomegalovirus-specific CD8 T cell responses. J Immunol. 2005;175:6123–6132. doi: 10.4049/jimmunol.175.9.6123. [DOI] [PubMed] [Google Scholar]
- 34.Lehner P.J., Wang E.C., Moss P.A., Williams S., Platt K., Friedman S.M., Bell J.I., Borysiewicz L.K. Human HLA-A0201-restricted cytotoxic T lymphocyte recognition of influenza A is dominated by T cells bearing the V beta 17 gene segment. J Exp Med. 1995;181:79–91. doi: 10.1084/jem.181.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benati D., Galperin M., Lambotte O., Gras S., Lim A., Mukhopadhyay M., Nouel A., Campbell K.A., Lemercier B., Claireaux M., Hendou S., Lechat P., de Truchis P., Boufassa F., Rossjohn J., Delfraissy J.F., Arenzana-Seisdedos F., Chakrabarti L.A. Public T cell receptors confer high-avidity CD4 responses to HIV controllers. J Clin Invest. 2016;126:2093–2108. doi: 10.1172/JCI83792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galperin M., Farenc C., Mukhopadhyay M., Jayasinghe D., Decroos A., Benati D., Tan L.L., Ciacchi L., Reid H.H., Rossjohn J., Chakrabarti L.A., Gras S. CD4(+) T cell-mediated HLA class II cross-restriction in HIV controllers. Sci Immunol. 2018;3 doi: 10.1126/sciimmunol.aat0687. [DOI] [PubMed] [Google Scholar]
- 37.Risnes L.F., Christophersen A., Dahal-Koirala S., Neumann R.S., Sandve G.K., Sarna V.K., Lundin K.E., Qiao S.W., Sollid L.M. Disease-driving CD4+ T cell clonotypes persist for decades in celiac disease. J Clin Invest. 2018;128:2642–2650. doi: 10.1172/JCI98819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Britanova O.V., Putintseva E.V., Shugay M., Merzlyak E.M., Turchaninova M.A., Staroverov D.B., Bolotin D.A., Lukyanov S., Bogdanova E.A., Mamedov I.Z., Lebedev Y.B., Chudakov D.M. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J Immunol. 2014;192:2689–2698. doi: 10.4049/jimmunol.1302064. [DOI] [PubMed] [Google Scholar]
- 39.Ge X., Tan V., Bollyky P.L., Standifer N.E., James E.A., Kwok W.W. Assessment of seasonal influenza A virus-specific CD4 T-cell responses to 2009 pandemic H1N1 swine-origin influenza A virus. J Virol. 2010;84:3312–3319. doi: 10.1128/JVI.02226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boria I., Cotella D., Dianzani I., Santoro C., Sblattero D. Primer sets for cloning the human repertoire of T cell receptor variable regions. BMC Immunol. 2008;9:50. doi: 10.1186/1471-2172-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dash P., Wang G.C., Thomas P.G. Single-cell analysis of T-cell receptor alphabeta repertoire. Methods Mol Biol. 2015;1343:181–197. doi: 10.1007/978-1-4939-2963-4_15. [DOI] [PubMed] [Google Scholar]
- 42.Brochet X., Lefranc M.P., Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giudicelli V., Brochet X., Lefranc M.P. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb Protoc. 2011;2011:695–715. doi: 10.1101/pdb.prot5633. [DOI] [PubMed] [Google Scholar]