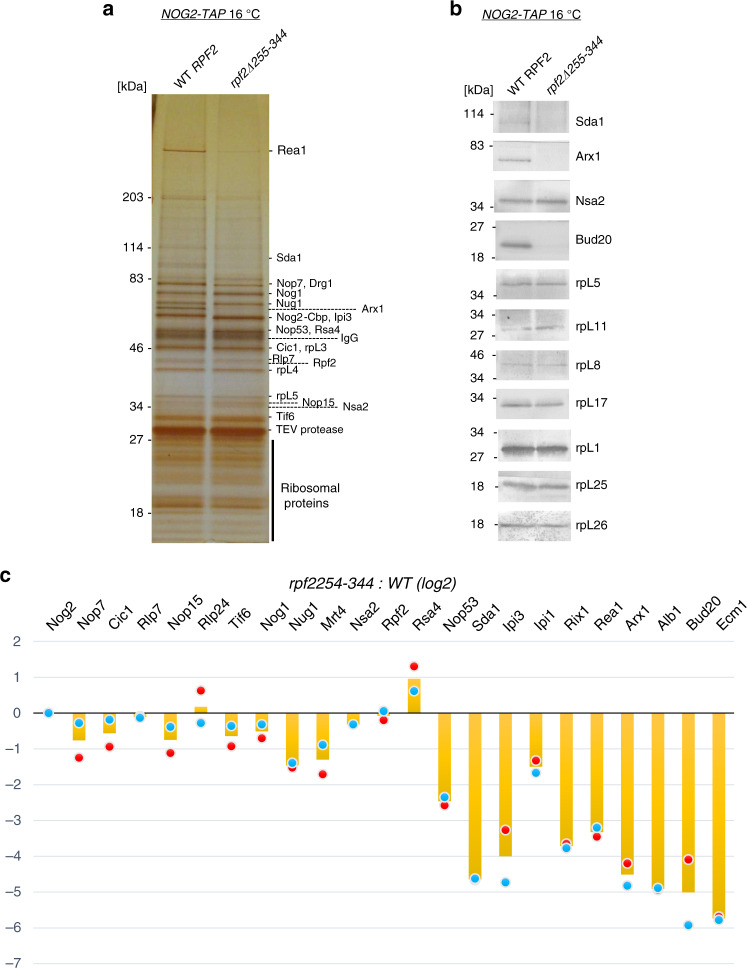
Fig. 2. AFs necessary for 5S rotation and export are affected in the rpf2Δ255-344 mutant.
a SDS-PAGE of proteins in assembling 60S subunits from wild-type cells or from the rpf2Δ255-344 mutant shifted to 16 °C for 5 h. Pre-ribosomal particles were purified using AF Nog2 as a bait and protein constituents were separated by electrophoresis and stained with silver. Molecular weight standards, the Nog2 bait protein Nog2-Cbp (calmodulin-binding peptide left behind after TEV cleavage), and relevant AFs are labeled. b Samples from a were subjected to western blotting using antibodies against specific proteins. All samples were derived from the same experiment and western blottings were processed in parallel. c Samples prepared as described in a were used for iTRAQ (semi-quantitative mass spectrometry) to quantify relative changes in levels of AFs in the genomic rpf2Δ255-344 mutant compared with the wild-type strain. The ratios were normalized to levels of Nog2 (bait). The fold change is shown using bar graphs (orange) in log2 scale as an average of two biologically independent samples (n = 2) for the genomic rpf2Δ255-344 mutant compared with the wild-type grown at 16 °C. Dot blots represent values for each biological replicate (red and blue). Source data are provided as a Source Data file.