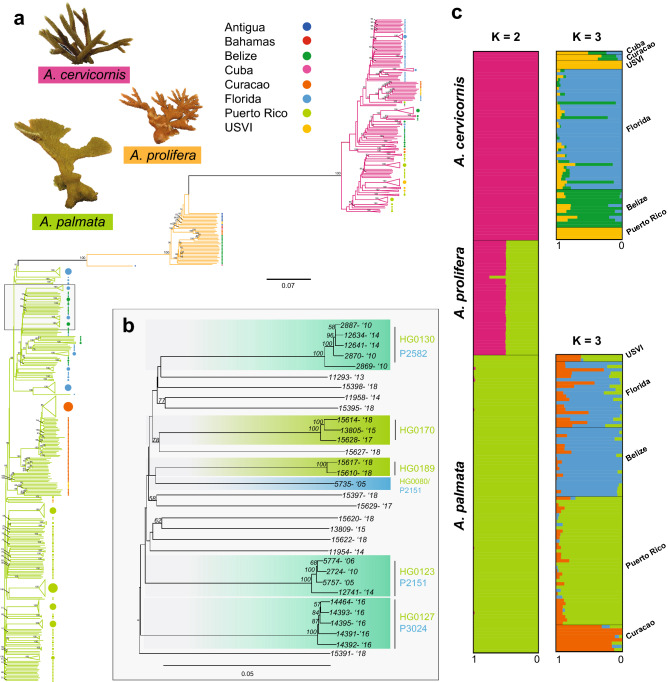
Figure 4.
Caribbean acroporid population analysis. Prevosti’s genetic distance of 19,694 SNPs was used to construct a neighbor-joining tree (a). The branches are colored by their genetic species identification and collection locations are indicated by the color of the circle at the terminal ends (Antigua = blue, Bahamas = red, Belize = green, Cuba = pink, Curacao = orange, Florida = light blue, Puerto Rico = light green, and USVI = yellow). Nodal support is based on the 100 bootstrap replicates. The nodes of genets with multiple ramets identified with the SNP data are collapsed in the tree. An example of genet resolution is provided based on the array SNP data and the previous microsatellite IDs over different collection years (b). The SNP genet ID is presented in green on the top and the microsatellite genet ID is presented in blue on the bottom. The clades are shaded blue-green where the two genotyping methods are congruent. The collection year is presented next to the sample identification number (- ‘xx). ADMIXTURE was run on a representative sample for each genet (n = 193), excluding genome samples, offspring of a Curacao cross and Puerto Rico samples from plate 9SR22844 (c). Individual bars represent the relative proportion of membership of a sample to the inferred K populations. Results from two source populations for all samples and three source populations for each species separately (K = 3 had the lowest cross-validation error for both species, Fig. S4).