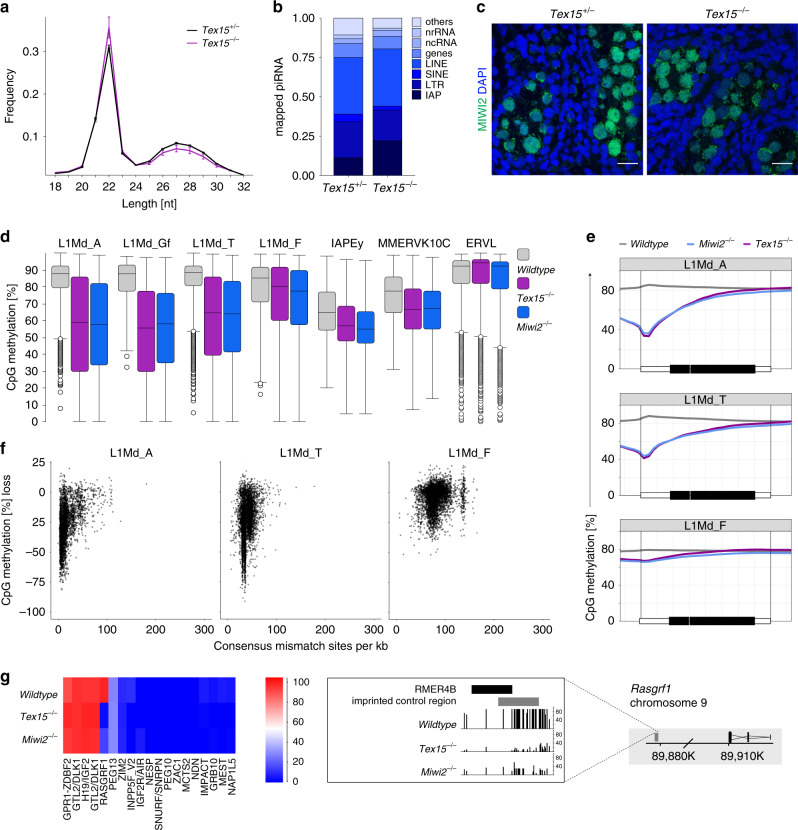
Fig. 3. TEX15 is essential for piRNA-directed de novo genome methylation.
a, b piRNAs analysis of Tex15+/− and Tex15−/− from small RNA sequenced E16.5 testes (n = 3). a Length distribution of small RNAs, data shows mean and s.e.m. P = 1, two sample t-test, Bonferroni adjustment. b piRNA annotation. c Representative images of MIWI2 stained E16.5 testis sections from Tex15+/− and Tex15−/− mice (n = 3). Scale bar 10 µm. d–g Genomic CpG methylation analysis from Wildtype, Tex15−/− and Miwi2−/− (n = 3) P14 undifferentiated spermatogonia. d CpG methylation percentage of TEs (non-overlapping genes). Box range indicates 25th to 75th percentile (interquantile range), line the median, whiskers range of median ± 2 interquantile range, dots datapoints outside this range. e Metaplots showing mean CpG methylation over LINE1 elements and adjacent 2 kb. f CpG methylation loss in Tex15−/− relative to Wildtype plotted against divergence from consensus sequence. g Mean CpG methylation level of maternal and paternal imprinted regions, Rasgrf1 shown in detail.