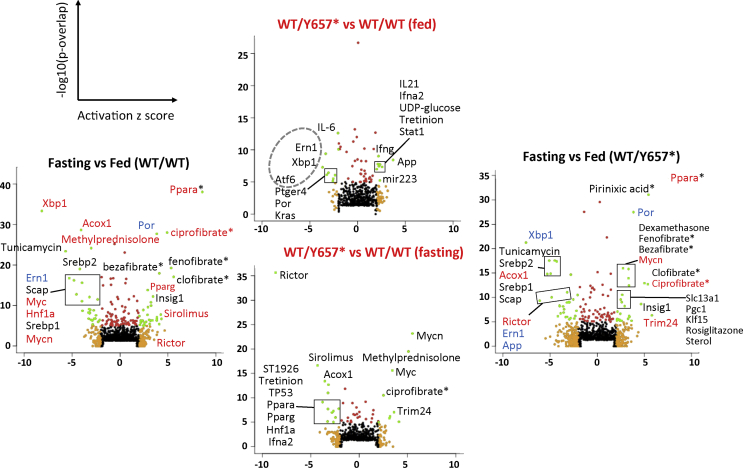
Figure 5.
Pathway analysis of liver transcriptomes of fed and fasted mice. Volcano plots are shown for predicted upstream regulators derived from Ingenuity® Pathway Analysis (IPA®) of transcriptomes of fed and fasted male Pik3r1WT/Y657∗ (n = 6,6) mice and wild-type littermates (n = 6,6). Outside plots show regulators showing differential activity in the fasting state compared to the fed state for wild-type (WT; left) and Pik3r1WT/Y657∗ (right) mice. Central plots show regulators with differential activity in Pik3r1WT/Y657∗ vs WT mice in the fed state (top) and fasting state (bottom). The green dots represent data point with an activation z score <−2 or >+2 and a p-value <1 × 10−5. Regulators showing differential activity in either genotype-based comparison are coloured red (activation z score > +2 in Pik3r1WT/Y657∗ vs WT mice) or blue (activation z score < −2 in Pik3r1WT/Y657∗ vs WT mice) in the outside plots showing differences based on nutritional state. Statistical analysis was performed using a general linear model with Bonferroni correction.