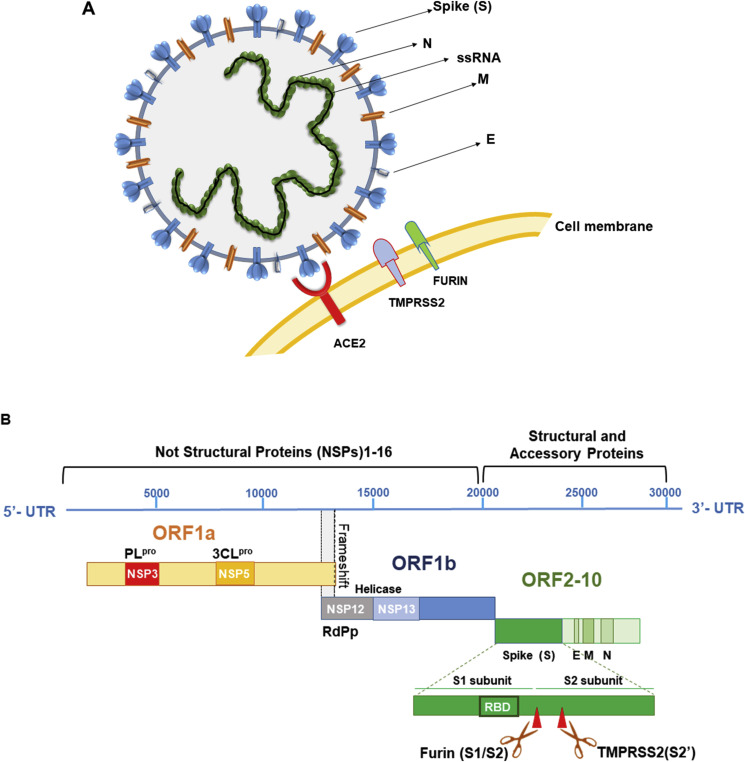
Fig. 2.
A. Coronaviruses form enveloped and spherical particles of 100–160 nm in diameter. They contain a positive-sense, single-stranded RNA (ssRNA) genome and nucleocapside proteins (N) that bind to RNA genome forming the nucleocapsid. The trimeric Spike glycoprotein (S) localizes on the surface of virus envelope and is essential for virus entry into the host cells. It recognizes the host receptor protein ACE2 on cell membrane after cleavage and activation by two host serine-proteases: TMPRSS2 and FURIN. Membrane or matrix protein (M) and small envelope protein (E) are both essential for the assembly and release of virions. B. SARS-CoV-2 genome, genes and proteins. There are 10 open reading frames (ORFs). The first ORF (67% of the genome) encodes not structural proteins (NSP), while the remaining ORFs give rise to accessory and structural proteins. ORF1a/b translates two polyproteins: pp1a and pp1b for the presence of a frameshift between ORF1a and ORF1b. These polyproteins are processed by a main protease known as 3C-like-protease (3CLpro) and one or two papain–like proteases (PLpro) into 16 NSPs. NSPs produce replicase complex essential for viral replication: NSP12 encodes RNA dependent RNA Polimerase (RdPd) and NSP13 encodes Helicase. ORFs 2–10 encode viral structural proteins: Spike (S), Envelope (E), Membrane (M), Nucleocapsid (N) and other auxiliary proteins. In particular, Spike protein comprises two regions: S1 with the receptor-binding domain (RBD) essential for the recognition of host receptor and S2, essential for membrane fusion and entry. Between S1 and S2 subunits there is the polybasic sequence recognized by host endo-proteases Furin. The activation site of S protein, is recognized by serine protease TMPRSS2 in region S2′ of S2 domain.