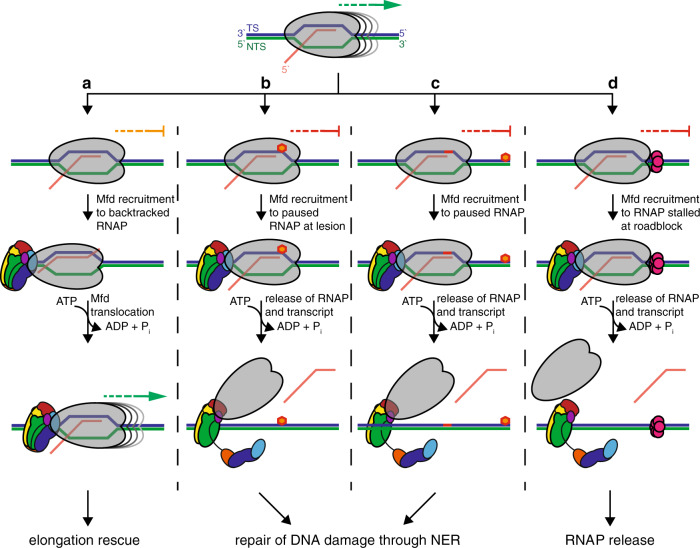
Fig. 1. All Mfd functions are dependent on translocation on DNA or action upstream of transcription elongation complexes.
Shown at the top is a schematic of an elongating RNAP (gray) with labeled nucleic-acid moieties (RNA, red; NTS, non-template strand, green; TS, template strand, blue) that can either become stalled or temporarily paused as indicated in a–d. a In Mfd-dependent rescue of class II transcriptional pausing, Mfd binds to backtracked RNAPs and promotes their forward translocation thereby rescuing transcript elongation16,26. b In canonical TCR, RNAP becomes stalled at a lesion (orange hexagon) on the TS and recruits Mfd (colored by domain, with D1a and D1b in blue, D2 in cyan, D3 in orange, D4 (RID) in magenta, D5 in yellow, D6 in green and D7 in red), either through 3D-diffusion or 1D-diffusion, enabled by a catch-up-then release mechanism dependent on ATP hydrolysis16, disruption of the D2–D7 clamp, RNA release, and UvrAB recruitment29,31. c In repair “at a distance”, Mfd is recruited to RNAP paused at class II pause signals (red line), which is first released off the nucleic-acid chains and acts as a processivity factor for Mfd to translocate toward downstream lesions in the TS, and initiate strand-specific repair17. d When RNAPs collides with protein roadblocks head-on, including replisomes, Mfd will release RNAP off the nucleic-acid chains, thereby freeing the DNA for other DNA-based processes18,19.