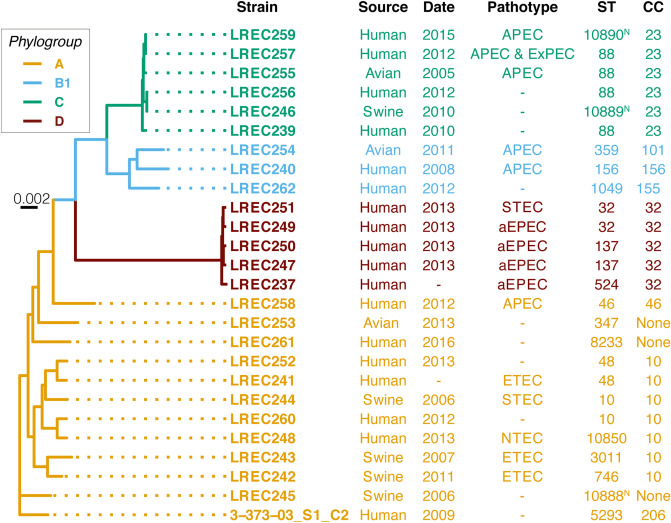
Figure 1.
Maximum-likelihood tree generated from the core-genome data of new pipolin-harboring strains. Strain names are colored according to the phylogenetic groups as indicated. Previously described pipolin-harboring isolate 3-373-03_S1_C2 was included as a reference. The best-fit model was GTR + F + R2 for all considered criteria in ModelFinder62. Scale bar indicates substitution rate per site. The main features are indicated on the right: source, isolation date, pathotype, sequence type (ST) and clonal complex (CC). New ST combinations assigned at Enterobase are indicated with N.