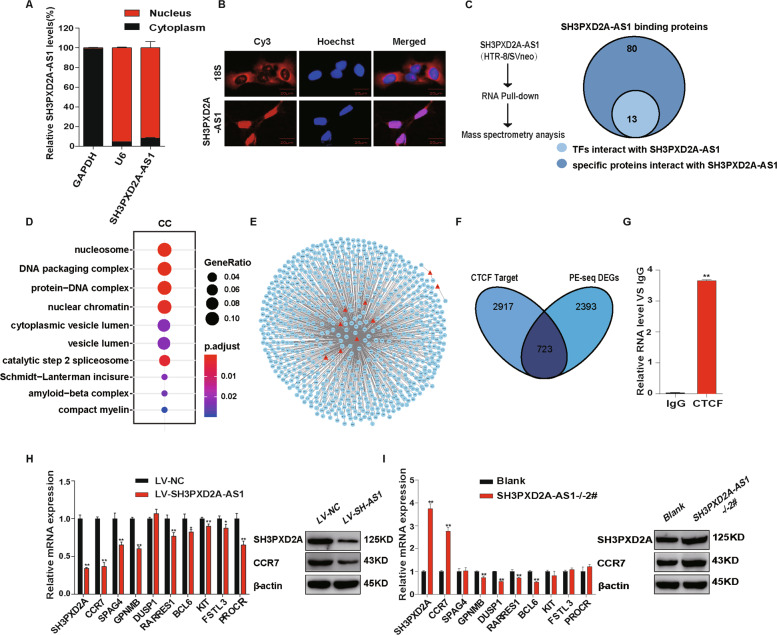
Fig. 4. SH3PXD2A-AS1 binds to transcription factors and regulates genes potentially involved in EOSPE.
a The subcellular localization of SH3PXD2A-AS1 in HTR-8/SVneo cell detected using cell fractionation. U6: nucleus marker; GAPDH: cytoplasm marker. b The subcellular localization of SH3PXD2A-AS1 in HTR-8/SVneo detected using FISH. Red: SH3PXD2A-AS1; blue: nucleus. c Proteins interacting with SH3PXD2A-AS1 detected in HTR8/SVneo cell by pulldown and mass spectrometry. d GO terms (Cellular Components) with over-presentation of SH3PXD2A-AS1-binding proteins. e TF-target network for DEG of EOSPE. Red triangles represent transcription factors (TFs) binding SH3PXD2A-AS1; blue circles represent differentially expressed target genes in EOSPE; f overlaps between CTCF target genes (3640) curated in database and differentially expressed genes (3116) detected in the transcriptome sequencing on placentae of patients with EOSPE. g Binding proteins of SH3PXD2A-AS1 detected by RIP assays and qRT-PCR. h SH3PXD2A-AS1 overexpression reduces CTCF target genes. Top 10 target genes with highest expression change in transcriptomes of EOSPE patients were evaluated using qRT-PCR. Left histogram blot shows the results of qRT-PCR. The reduced expression of two targets were verified by western blot (left). i SH3PXD2A-AS1 knockout increases CTCF target genes. Top 10 target genes with highest expression change in transcriptomes of EOSPE patients were evaluated using qRT-PCR. Left histogram blot shows the results of qRT-PCR. The increased expression of two targets were verified by western blot (left). **p < 0.01, *p < 0.05.