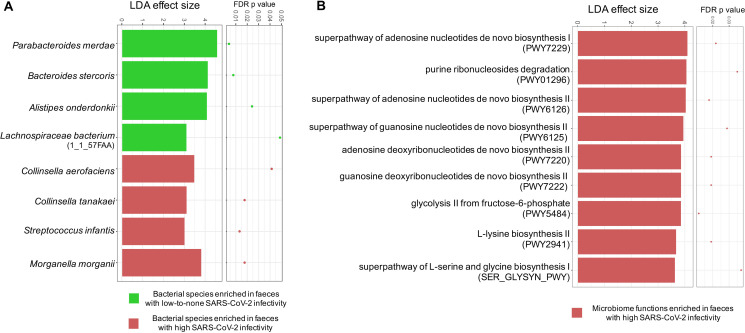
Figure 4.
Differential bacterial species and functional capacities between faeces with high severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectivity and faeces with low-to-none SARS-CoV-2 infectivity. Differential bacterial species (A) and functionality (B) were identified via LefSE analysis across all time-point stools of 15 patients with COVID-19. Only species and functional modules with LDA effect size >2 and FDR-corrected p value <0.05 were plotted. High SARS-CoV-2 infectivity was defined as higher 3’ vs 5’ end coverage of SARS-CoV-2 genome in faecal viral RNA metagenome. Low-to-none SARS-CoV-2 infectivity was defined as similar 3’ and 5’ end coverage or no coverage of the SARS-CoV-2 genome in faecal viral RNA metagenome.