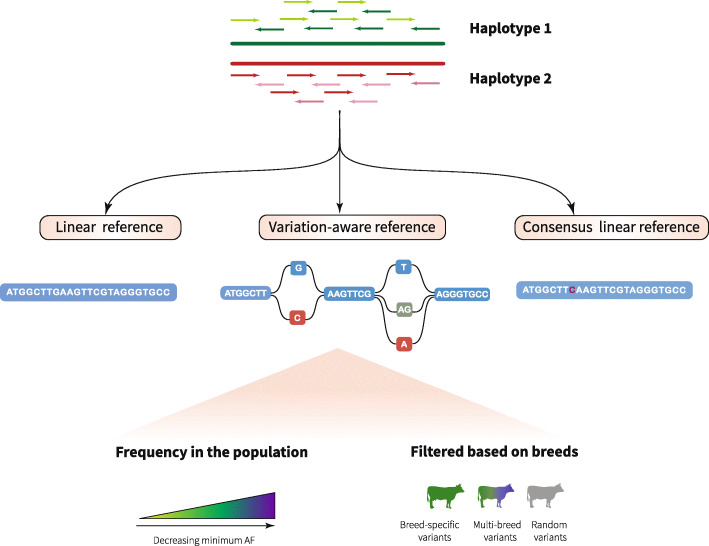
Fig. 1.
Schematic overview of the construction of breed-specific augmented genome graphs. We used the vg toolkit to augment the bovine linear reference sequence (ARS-UCD1.2) with alleles at SNPs and Indels that were discovered in 288 cattle from four breeds. Alleles that were added to the linear reference were prioritized based on their alternate allele frequency (AF). Reads simulated from true haplotypes were aligned to variation-aware, linear and consensus reference sequences to assess read mapping accuracy on cattle chromosome 25. Short-read sequencing data of Brown Swiss cattle were used to investigate sequence variant genotyping accuracy and reference allele bias using a bovine whole-genome graph as a novel reference