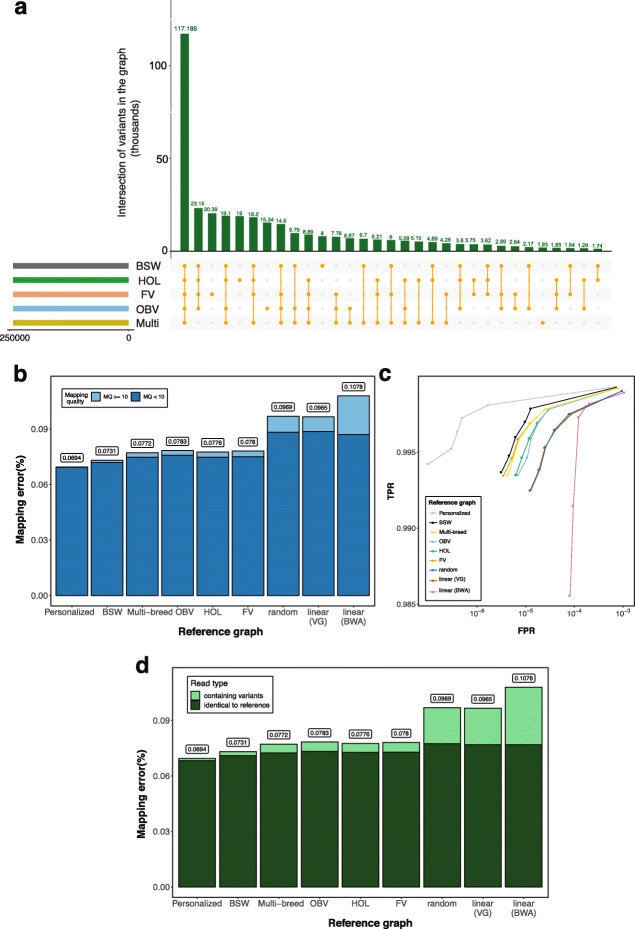
Fig. 4.
The accuracy of mapping simulated BSW paired-end reads to variation-aware and linear reference structures. a We added 243,145 chromosome 25 variants to the Hereford-based reference sequence that were filtered for alternate allele frequency > 0.03 in either the BSW, FV, HOL, or OBV populations. The pan-genome graph (Multi) contained 243,145 variants that had alternate allele frequency threshold > 0.03 across 288 cattle from the four breeds considered. The bars indicate the overlap of variants (averaged across ten replications) that were added to different graphs. b Proportion of simulated BSW reads that mapped erroneously against personalized graphs, breed-specific augmented graphs, pan-genome graphs (Multi-breed), random graphs, or linear reference sequences. We used vg and BWA mem for linear mapping. Dark and light blue colors represent the proportion of incorrectly mapped reads that had phred-scaled mapping quality (MQ) < 10 and MQ > 10, respectively. c True-positive (sensitivity) and false-positive mapping rate (specificity) parameterized on mapping quality. d Proportion of BSW reads that mapped incorrectly against breed-specific augmented graphs, pan-genome graphs (Multi-breed), random graphs, or linear reference sequences. Dark and light green colors represent the proportion of incorrectly mapped reads that matched corresponding reference nucleotides and contained non-reference alleles, respectively. Results for single-end mapping are presented in Additional file 1: Fig. S7