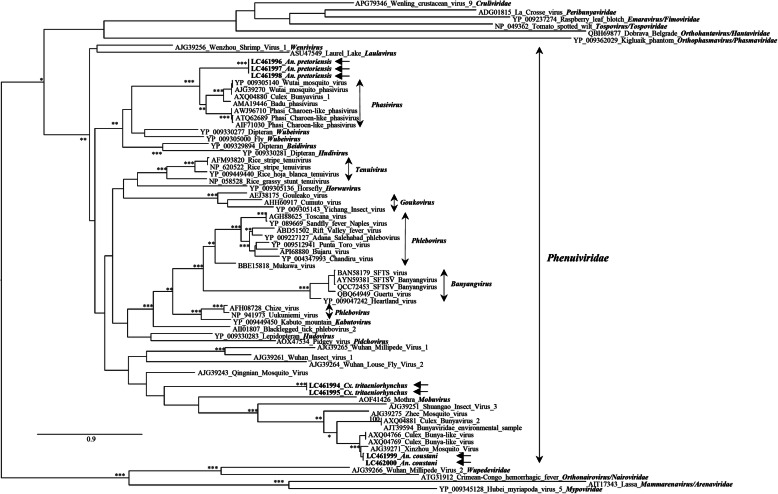
Fig. 3.
a Phylogenetic analysis of partial flavivirus NS5 nucleotide sequences (0.5 kbp DNA amplicon) from insect-specific flaviviruses. cISF and dISF indicate classical and dual-host associated insect-specific flaviviruses, respectively. At specific branches, the number of “*” indicates the support revealed by the different phylogenetic reconstructions methods used (bootstrap values ≥75% and posterior probability values ≥0.80 were assumed as significant). The sequences described in this work are indicated in bold-face, and their origin (mosquito species) is also indicated. All the sequences used are designated by their respective accession numbers|virus name. The size bar indicates the number of nucleotide substitutions per site. b amplification of flavivirus-like from RNA and mosquito genomic DNA using a combination of treatments that included the use/or not (+/) of DNase I followed/or not (+/−) by reverse transcription (RT). The NZYtech ladder VI was used as a molecular mass marker. c Structure of the flavivirus wild-type and the NS5 fragments analyzed. The 3′-end of the viral genome is schematically shown at the top. §-Mosquito species could not be confirmed by COI sequence