Figure 1. Constitutive PRDM1 expression driven by PAX6-CRE does not prevent progenitor VSX2 expression.
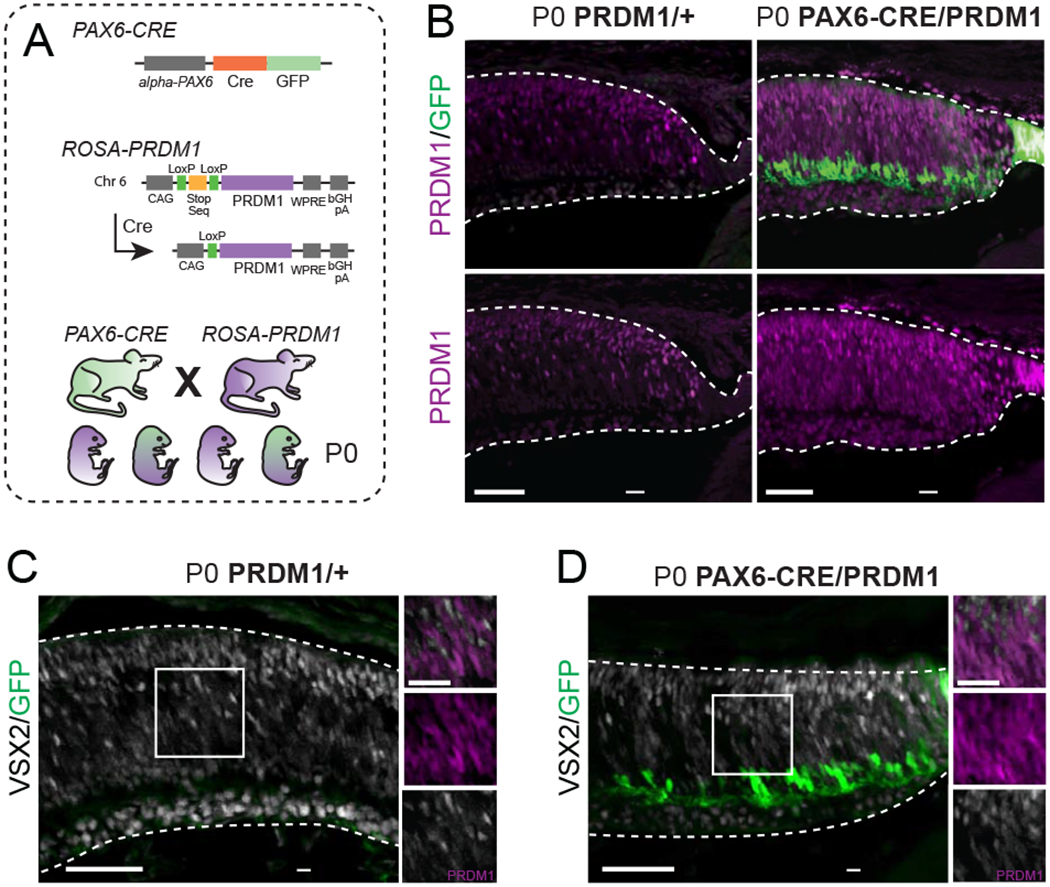
A) Schematic of transgenic mice utilized. To make the PRDM1 strain, the ROSA26 locus on chromosome 6 was targeted to insert the Prdm1 cDNA downstream of a CAG enhancer and a LoxP-stop-LoxP sequence. Further downstream is a woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) and a bovine growth hormone polyadenylation (bGH pA) site. Introduction of CRE removes the stop sequence and allows for permanent PRDM1 expression. B) Representative immunohistochemistry of PRDM1/+ control compared to PAX6-CRE/PRDM1 retinas stained for Prdm1 (purple) and GFP (green). Note the ectopic expression of Prdm1 in the RPE and ciliary body of PAX6-CRE/PRDM1 eyes (arrowheads). C-D) P0 PRDM1/+ control compared to PAX6-CRE/PRDM1 retinas stained for VSX2 (white), GFP (green), and PRDM1 (purple, inset-only). Arrowheads mark PRDM1+/VSX2+ cells. There is VSX2 background staining in the ganglion cell layer in both conditions. bars=50μm, inset bars=25μm.
