Figure 1.
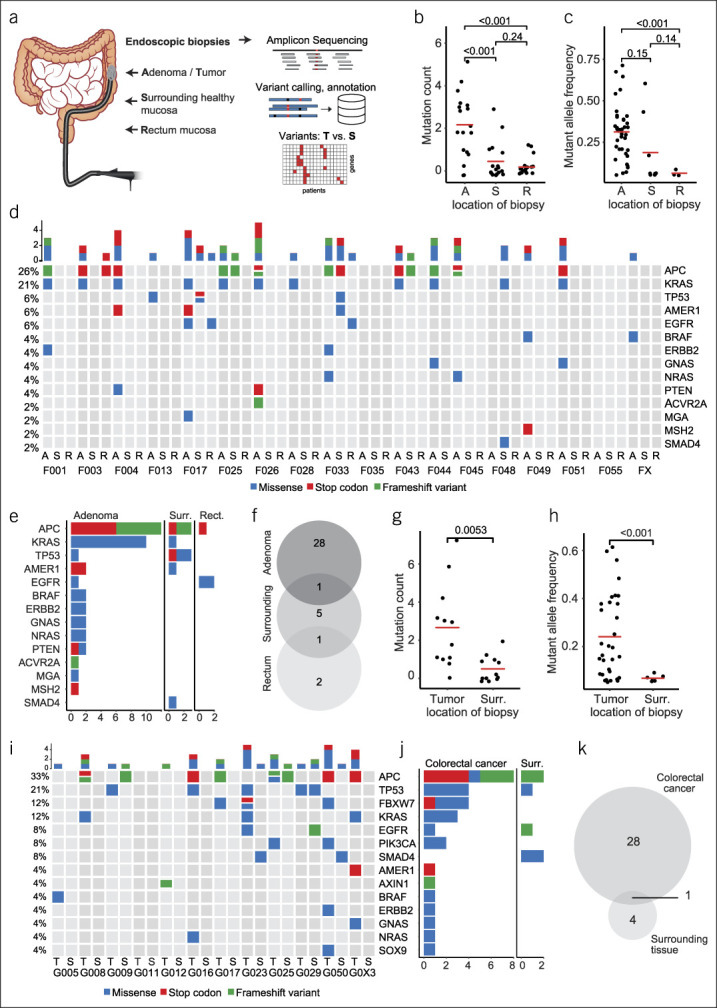
Systematic mutational profiling of colorectal neoplasms and adjacent normal colorectal mucosa. (a) Schematic representation of biopsy acquisition and experimental workflow. Patients with either large colorectal adenomas or colorectal cancer (CRC) were recruited. Biopsies were obtained using a forceps via colonoscopy from the neoplasia, adjacent normal mucosa, and in case of adenomas also from healthy rectum mucosa. DNA isolated from all samples underwent amplicon sequencing of frequently mutated loci in CRC-associated genes and subsequent bioinformatics analysis. (b) Mutation count in adenoma, surrounding mucosa and rectum mucosa samples. (c) Fraction of mutated alleles detected in adenoma, surrounding mucosa and rectum mucosa samples. (d) Oncoprint of all mutations identified in adenomas (A), surrounding mucosa (S) and rectal mucosa (R) from 20 patients. Additional rectal mucosa samples were not analyzed in 3 patients with rectal adenomas. (e) Frequency of mutations by cancer-gene stratified by location of biopsy (adenoma, surr., surrounding, rect., rectum) (f) Venn diagram showing the concordance of mutations in adenomas, surrounding healthy mucosa and rectal mucosa. (g) Mutation count in CRC and surrounding mucosa. (h) Fraction of mutated alleles detected in CRCs and surrounding mucosa. (i) Oncoprint of all mutations identified in CRCs (T = tumor) and surrounding mucosa (S = surrounding). (j) Frequency of mutations by cancer-gene, stratified by location of biopsy (CRC vs surr., surrounding) (k) Venn diagram showing the concordance of mutations in CRC and surrounding healthy mucosa.
