Figure 5. Identification of DNA-Binding Factors that Co-occupy Region-Specific Regulatory Elements.
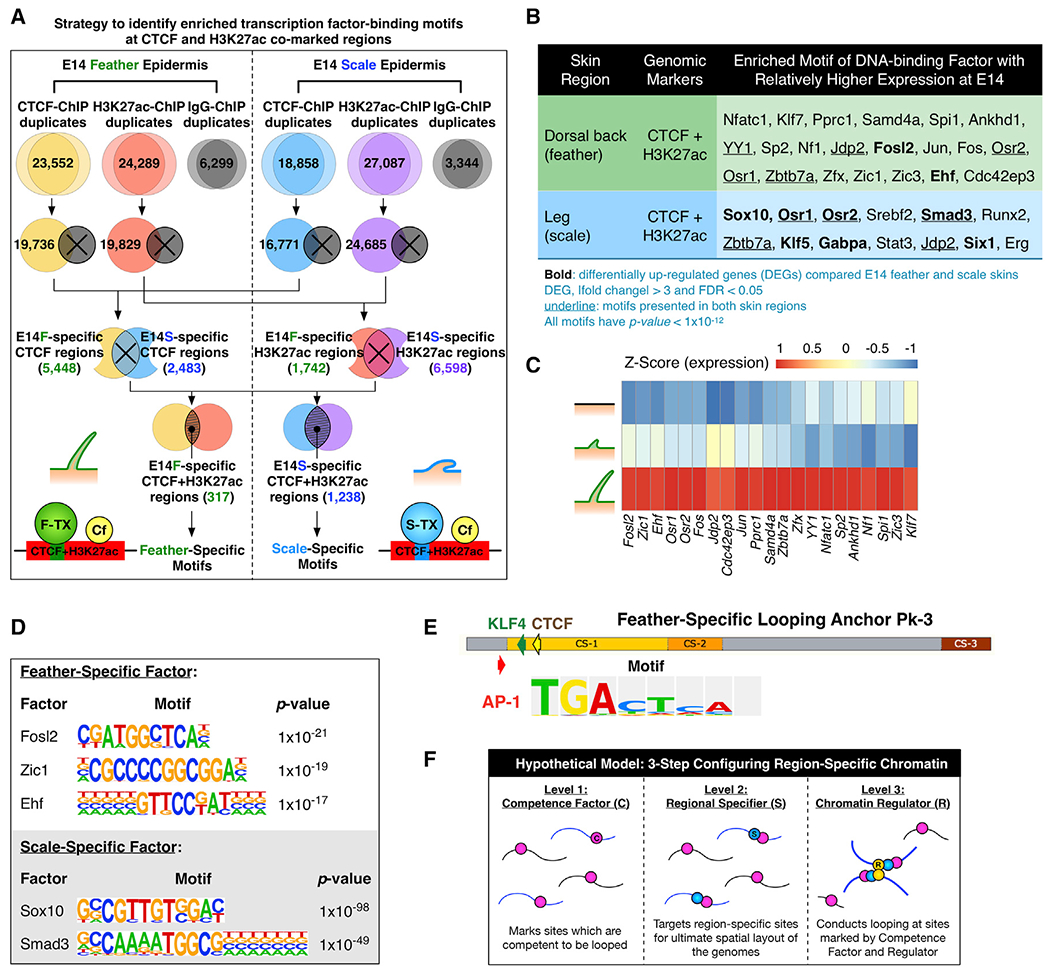
(A) Schematic depicting the analysis strategy used to identify motifs of region-specific transcription factors in CTCF/H3K27ac co-marked chromatin.
(B) HOMER de novo motif discovery showing enriched motifs of DNA-binding factors at CTCF and H3K27ac co-marked chromatin in E14 feathers and scales. Expression of the listed factors was validated using RNA-seq results.
(C) Normalized expression of skin region-specific DNA-binding factors during skin specification. Z score, [RPKM-(mean RPKM)/standard deviation]. The order of factors was sorted by differential expression.
(D) Lists of skin-region-specific binding motifs.
(E) Positions of an AP1 factor binding motif in feather-specific looping anchor Pk3 of the Chr27 β-krt cluster.
(F) The three-factor hypothesis elucidating how region-specific chromatin looping is established – competence factor, regional specifier, and chromatin regulators.
