Figure 4.
The SARS-CoV-2 nsp12-NiRAN Domain, Pseudokinase SelO, and ADP Binding
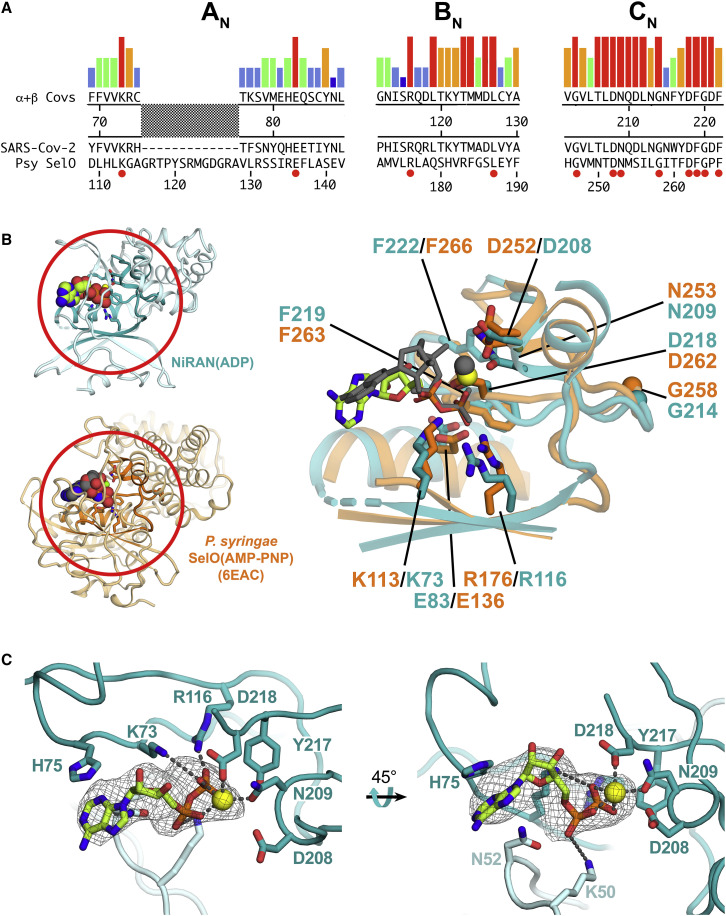
(A) The colored histograms denote identity in a sequence alignment of 45 α- and β-CoV nsp12 sequences (red bar, 100% identity; dark blue bar, 20% or less) in the N-terminal signature motifs AN, BN, and CN (Lehmann et al., 2015a) of the NiRAN domain. The consensus sequence is shown below. The SARS-CoV-2 nsp12 and the pseudokinase P. syringae (Psy) SelO (Sreelatha et al., 2018) are aligned below. Residues that are 100% identical in the nsp12 alignment and conserved in SelO are highlighted by a red dot underneath.
(B) Left: structures of the SARS-CoV-2 NiRAN domain (cyan ribbon) with ADP-Mg2+ (spheres) and Psy SelO (orange) with AMP-PNP-Mg2+ (PDB: 6EAC; Sreelatha et al., 2018). The AN, BN, and CN regions are highlighted. Right: structure-based alignment via α-carbons of the AN, BN, and CN regions, with side chains of conserved residues shown. The α- and β-phosphates of the NiRAN domain ADP-Mg2+ (lime carbon atoms and yellow sphere, respectively) superimpose almost exactly with the β- and γ-phosphates of the SelO AMP-PNP-Mg2+ (dark gray), whereas the nucleoside moieties diverge.
(C) Two views of the ADP-Mg2+-bound pocket of the SARS-CoV-2 NiRAN domain. Side chains interacting with the ADP-Mg2+ are shown (polar interactions are denoted by gray dashed lines). D208 likely makes a water-mediated interaction with Mg2+ (Sreelatha et al., 2018). The cryo-EM difference density for ADP-Mg2+ is shown (light gray mesh).
See also Data S3.