Figure 5.
Correspondence of Structural Determinants for Backtracking between Cellular Multi-subunit DdRp and SARS-CoV-2 RdRp
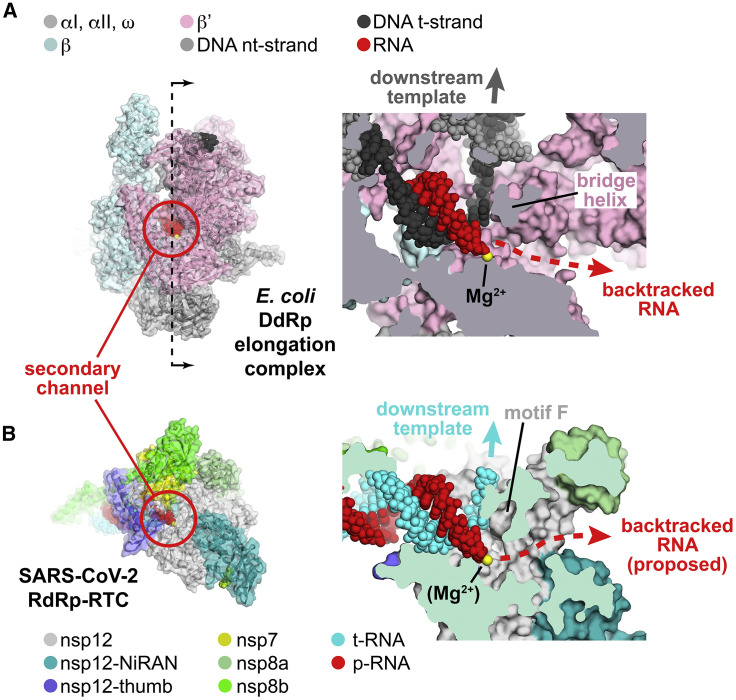
(A and B) Left: overall views of complexes. Proteins are shown as cartoon ribbons with transparent molecular surfaces. Nucleic acids are shown as atomic spheres. Color coding is shown in the keys. Right: cross-sectional view of the active site region (at the 3′ end of the RNA product).
(A) Left: structure of the E. coli DdRp transcription elongation complex (EC; Kang et al., 2017), viewed down the secondary channel. The DdRp active-site Mg2+-ion is shown as a yellow sphere. The secondary channel is highlighted in the red circle. The thin dashed line illustrated the cut and viewing direction of the cross-section on the right. Right: cross-sectional view showing the RNA/DNA hybrid. The bridge helix (viewed end-on in the cross-section) is denoted. The bridge helix directs the downstream template duplex DNA to the top (dark gray arrow). Under the bridge helix, the secondary channel allows NTP substrates to diffuse into the active site (Westover et al., 2004; Zhang et al., 1999) and accommodates the single-strand RNA transcript 3′ end in backtracked complexes (Abdelkareem et al., 2019; Cheung and Cramer, 2011; Wang et al., 2009).
(B) Left: view down the newly described secondary channel of the SARS-CoV-2 holo-RdRp. The secondary channel is highlighted in the red circle. Right: cross-sectional view showing the t-RNA/p-RNA hybrid. Motif F (viewed end-on in the cross-section) is denoted. Motif F directs the downstream t-RNA to the top (cyan arrow). Under motif F, the secondary channel could accommodate the single-strand p-RNA 3′ end in the event of backtracking.