Figure S6.
Comparison of the nsp132-RTC (CHAPSO) Structure with nsp131-RTC (CHAPSO) and nsp132-RTC (No Detergent), Related to Figures 2 and 3
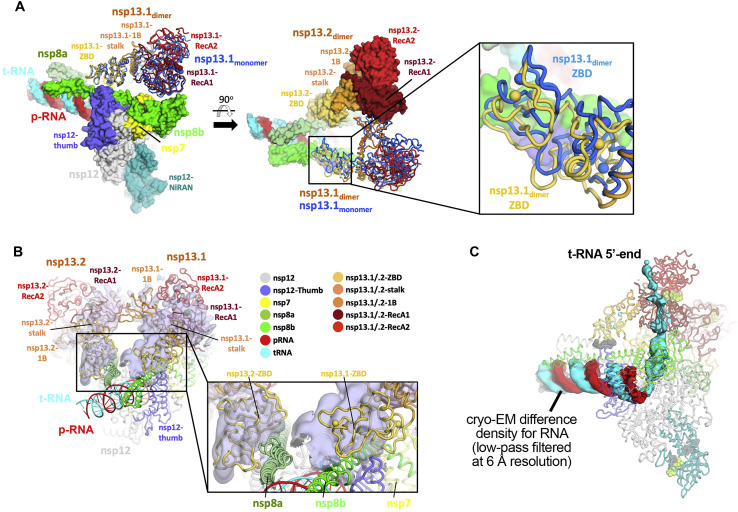
A. Structure of nsp132-RTC (CHAPSO) colored according to key in b and shown as a molecular surface except nsp13.1, which is shown as cartoon tubes. Superimposed on the overall structure is nsp13 (marine) modeled from the nsp131-RTC (CHAPSO). Overall RMSD (calculated using ‘rms_cur’ in PyMOL) between the two nsp13.1 structures is 8.1 Å over 596 Cα atoms. (left) overall structure. (middle) overall structure rotated 90°. (right) zoom-in of boxed region in middle panel, showing region around nsp13.1-ZBD.s RMSD (calculated using ‘rms_cur’ in PyMOL) between the two nsp13.1 ZBDs is 3.6 Å over 100 Cα atoms.
B. Structure of nsp132-RTC (CHAPSO) is shown in cartoon tubes, colored based on key, and superimposed onto the cryo-EM map from the nsp132-RTC (no detergent) dataset (shown as light blue transparent surface). Density map is locally filtered by resolution and difference density for nsp13 is highlighted using ‘isosurf’ command in PyMOL with 10 Å carve buffer. (left) overall structure. (right) zoom-in of boxed region in left panel, showing region around nsp13-ZBDs.
C. Structure of nsp132-RTC (CHAPSO) colored according to key in (b), the view is similar to the view of Figure S6A(left). Protein is shown as pale, transparent backbone worms. The surface shows a cryo-EM different density for the RNA (t-RNA, cyan; p-RNA, red) low-pass filtered to 6 Å resolution. The difference map was generated by calculating a map from the nsp132-RTC coordinates with the RNA removed using the molmap command in Chimera (Pettersen et al., 2004), subtracting this map from the experimental map (Chimera vop command), then low-pass filtering this difference map at 6 Å resolution).