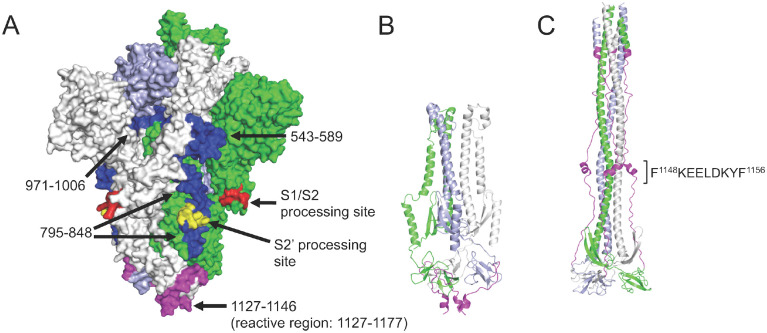
Figure 3. Recurrent Spike protein epitopes correspond to accessible and functionally-important sites within the protein structure.
A) Space-filling SARS-CoV-2 Spike protein structure (PDB id: 6VYB) (Walls et al., 2020) showing the native trimer (monomers shown in green, gray and light blue) with the four recurrent epitope regions targeted by COVID-19 convalescent IgG (Figure 2C) highlighted in blue or magenta. Each epitope is identified by its amino acid range within the S protein sequence (GenBank: YP_009724390.1). The epitope at positions 1127–1177 (magenta) includes a region that is unresolved in the structure (1147–1177). Protease processing sites are highlighted in red and yellow, including the S2’ site that occurs within the 795–848 epitope. B) Ribbon model of the SARS-CoV-2 Spike S2 subunit after protease processing, using the same color scheme as in (A). C) Ribbon model of the 6-helical bundle (post-fusion) conformation of the S2 subunit, built based on the Cryo-EM structure of mouse hepatitis virus (PDB id: 6B3O) (Walls et al., 2017). The 1127–1177 region is again highlighted in magenta (with the minimal epitope region shown), and a comparison with (B) shows the dramatic conformational rearrangement that occurs at this site.