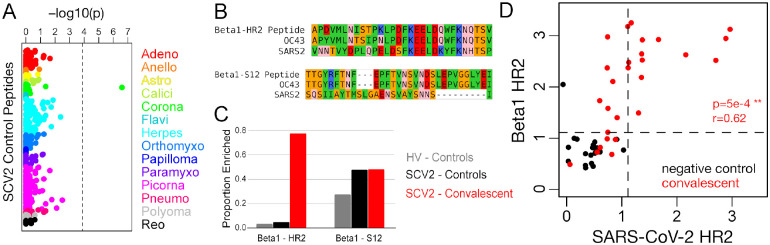
Figure 5. Spike HR2 antibodies elicited by SARS-CoV-2 strongly cross-react with the homologous region of Betacoronavirus 1.
A) Fisher’s exact test p-values measuring the correlation between donor SARS-CoV-2 status and enrichment for each of 373 control peptides. These peptides were designed from 55 virus species that belong to 14 different families (colors, labels correspond to family names with the omission of “-viridae”), and they recognize epitopes that we previously identified as commonly reactive in the general population. Dashed vertical line shows the Bonferroni-corrected threshold for significance. B) Sequence alignments between SARS-CoV-2 (SARS2) and the Betacoronavirus 1 (beta1-CoV) strain, hCoV-OC43 (OC43), at two Spike protein regions covered by SCV2 library control peptides designed from beta1-CoV (Beta1) sequences. Residues are colored according to amino acid properties: small non-polar (orange), hydrophobic (green), polar (pink), negatively charged (red) and positively charged (blue). C) Proportion of samples reactive to the two Betacoronavirus 1 peptides shown in panel (B). Two separate sets of negative controls are shown, those assayed with the HV peptide library (grey, n=33) and those assayed with the SCV2 peptide library (black, n=21). Results from COVID-19 convalescent samples are shown in red (n=27). D) Quantitative comparison of reactivities to homologous HR2 peptides from SARS-CoV-2 (x-axis) and beta1-CoV (y-axis) across the SCV2-characterized donor cohort. Axes represent log10(2 + Z-scores) and dashed lines indicate threshold for significance (Z-score ≥ 11).