Figure 1.
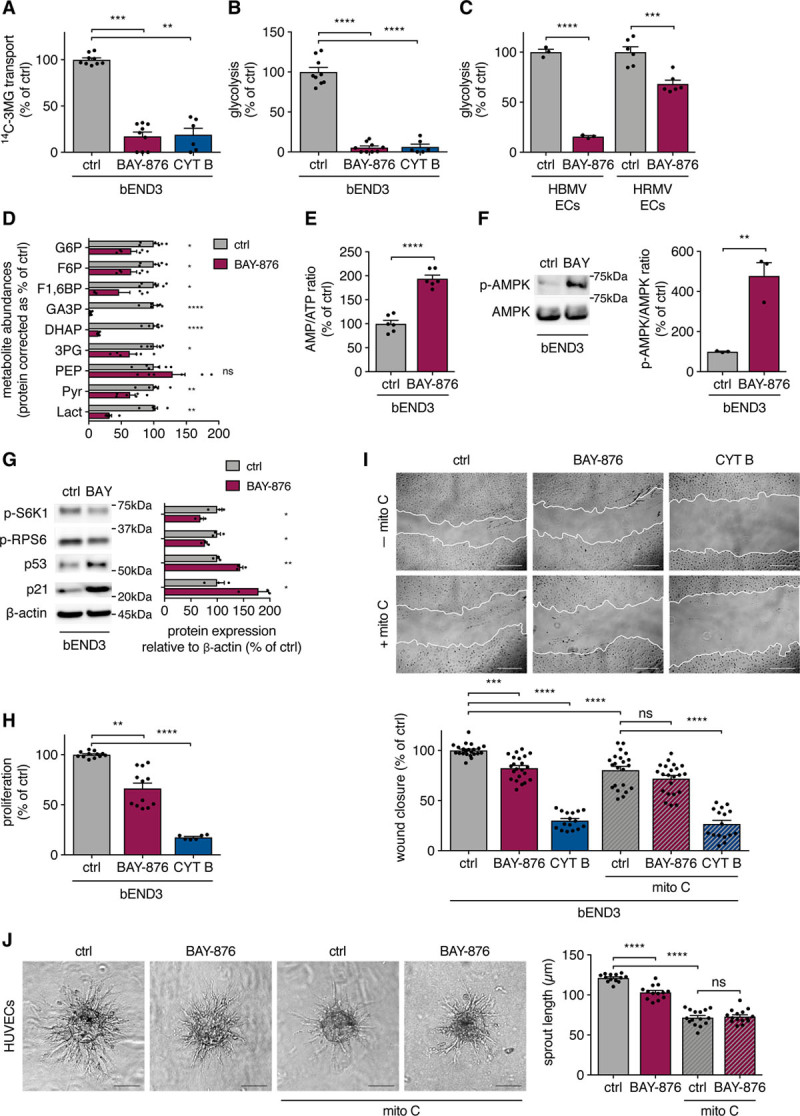
GLUT1 (glucose transporter isoform 1) inhibition impairs endothelial cell (EC) glucose metabolism and proliferation but not migration. A and B, 14C-3-O-methylglucose (3MG) transport (Kruskall-Wallis test and Dunn multiple comparisons test; A) and glycolytic flux (1-way ANOVA and Tukey multiple comparisons test; B) in cells from a brain-derived EC line (bEND3) incubated with 20 nmol/L BAY-876 (N4-[1-[(4-cyanophenyl)methyl]-5-methyl-3-(trifluoromethyl)-1H-pyrazol-4-yl]-7-fluoro-2,4-quinolinedicarboxamide) and 20 µmol/L cytochalasin B (CYT B) vs control. C, Glycolytic flux in human brain microvascular ECs (HBMV ECs) and human retinal microvascular ECs (HRMV ECs) incubated with 20 nmol/L BAY-876 vs control (Student t test). D, Abundances of glycolytic intermediates in bEND3 cells incubated with 20 nmol/L BAY-876 vs control (Student t test or Mann-Whitney U test). E, AMP/ATP ratio in cells from a brain-derived EC line (bEND3) incubated with 20 nmol/L BAY-876 vs control (Student t test). F and G, Western blot of p-AMPK (phospho-AMP-activated protein kinase), AMPK (F) and p-S6K1 (phospho-S6 kinase 1), p-RPS6 (phospho-ribosomal protein S6), p53, and p21 (G) in bEND3 cells incubated with 20 nmol/L BAY-876 vs control (Student t test). H, Proliferation rate of bEND3 cells incubated with 20 nmol/L BAY-876 and 20 µmol/L CYT B vs control (Kruskall-Wallis test and Dunn multiple comparisons test). I, Representative pictures and quantifications of scratch wound closure in bEND3 cells incubated with 20 nmol/L BAY-876 and 20 µmol/L CYT B vs control in conditions with and without mitomycin C (mito C) pretreatment (2-way ANOVA and Tukey multiple comparisons test). J, Representative pictures and quantifications of sprouting human umbilical vein EC (HUVEC) spheroids incubated with 40 nmol/L BAY-876 vs control in conditions with and without mitomycin C pretreatment (2-way ANOVA and Tukey multiple comparisons test). Scale bar=500 µm (I) and 100 µm (J). 3PG indicates 3-phosphoglycerate; AMP, adenosine monophosphate; ATP, adenosine triphosphate; DHAP, dihydroxyacetone phosphate; F1,6BP, fructose 1,6-bisphosphate; F6P, fructose 6-phosphate; G6P, glucose 6-phosphate; GA3P, glyceraldehyde 3-phosphate; Lact, lactate; PEP, phosphoenolpyruvate; and Pyr, pyruvate. *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001.
