Figure 6:
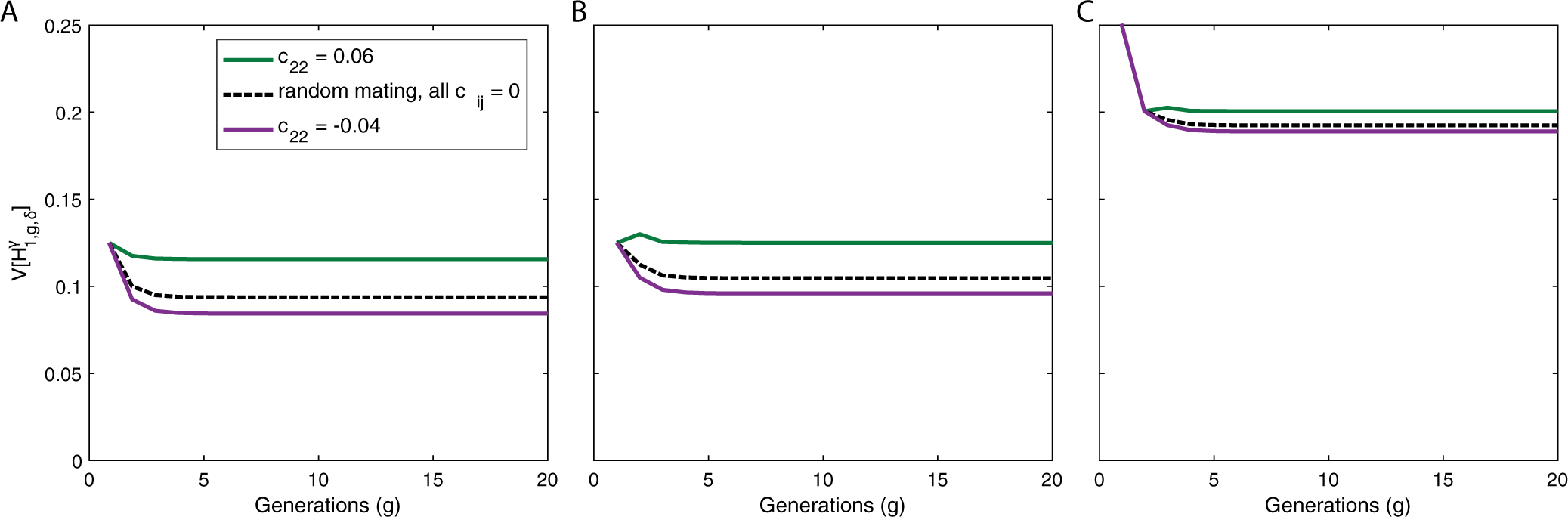
Example of the variance of ancestry under a constant-admixture scenario with assortative mating in producing g ≥ 2 when populations are allowed to differ in the direction of their mating preference. (A) Autosomes, . (B) X chromosomes in a female, . (C) X chromosomes in a male, . In all scenarios, populations S1 and H experience positive assortative mating, c11,chh = 0.02. Green curves show a scenario in which S2 also experiences positive assortative mating, c22 = 0.06; purple curves show a scenario in which S2 experiences negative assortative mating, c22 = −0.04. The scenario of random mating with the same contribution parameters is the black dashed line. The plots use eqs. (20), (21), and (29)–(32). We set , and use initial conditions and c11,0 = 0. In this example, negative assortative mating in S2 offsets the positive assortative mating in S1 and H, producing lower variances of admixture in S1, , in the assortatively mating population than in the randomly mating population.
