Figure 2. SMN genes generate a vast repertoire of circRNAs.
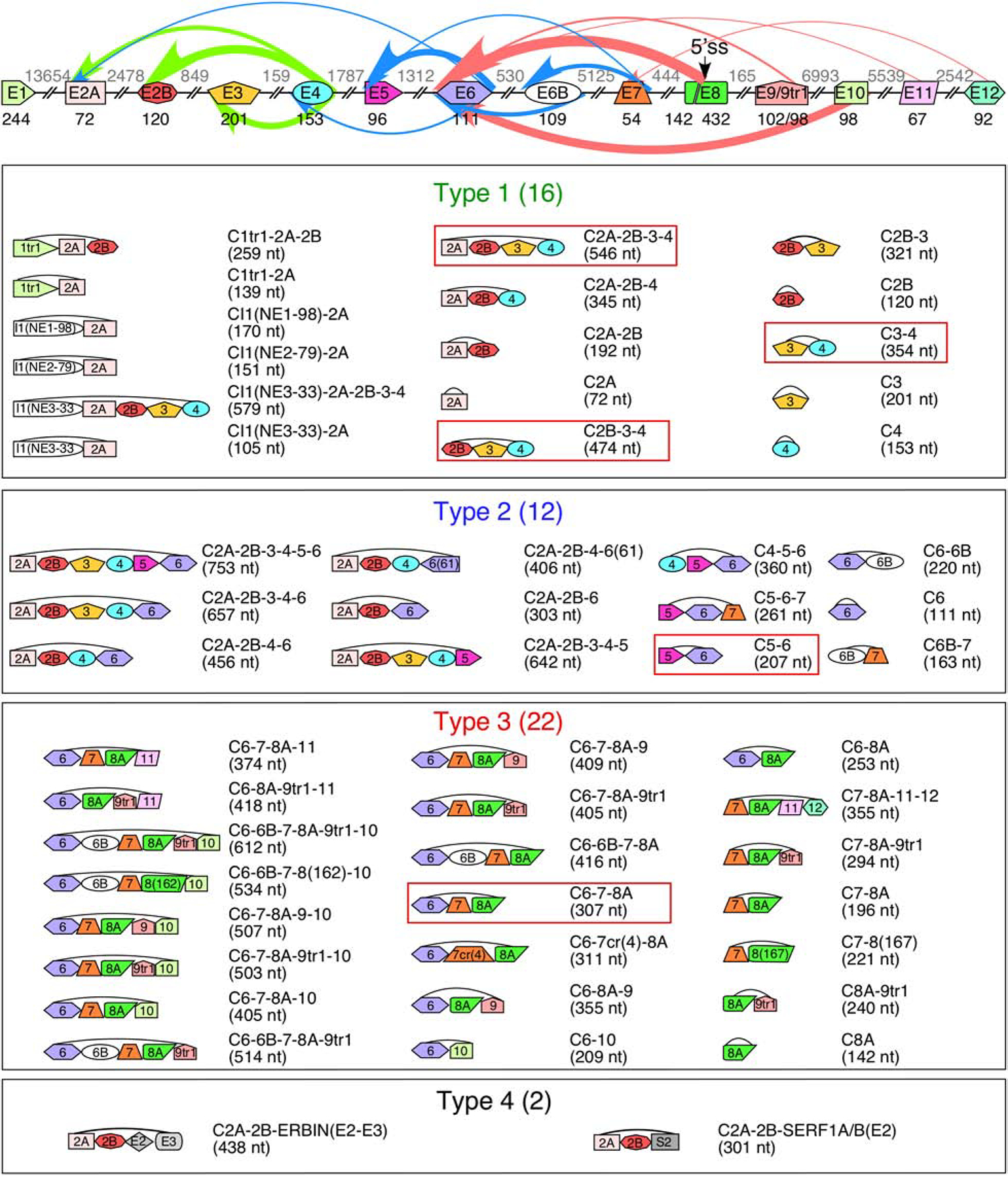
The genomic layout of the SMN genes and prominent backsplicing events is given at the top. Exons are shown as colored shapes, introns as broken lines. Exon sizes are given in black below each exon, intron sizes are given in gray above each intron. A cryptic 5′ss in exon 8 is indicated with an arrow. Colored arrows represent backsplicing events. The relative thickness of each arrow represents backsplice site usage frequency; arrow color represents type of circRNA (green for type 1, blue for type 2, and red for type 3). Bottom 4 panels show the total catalog of each type of SMN circRNA. The exon content of each circRNA is indicated graphically. The name of each circRNA is given at the right with total size in parentheses. circRNAs with the highest expression level are boxed.
