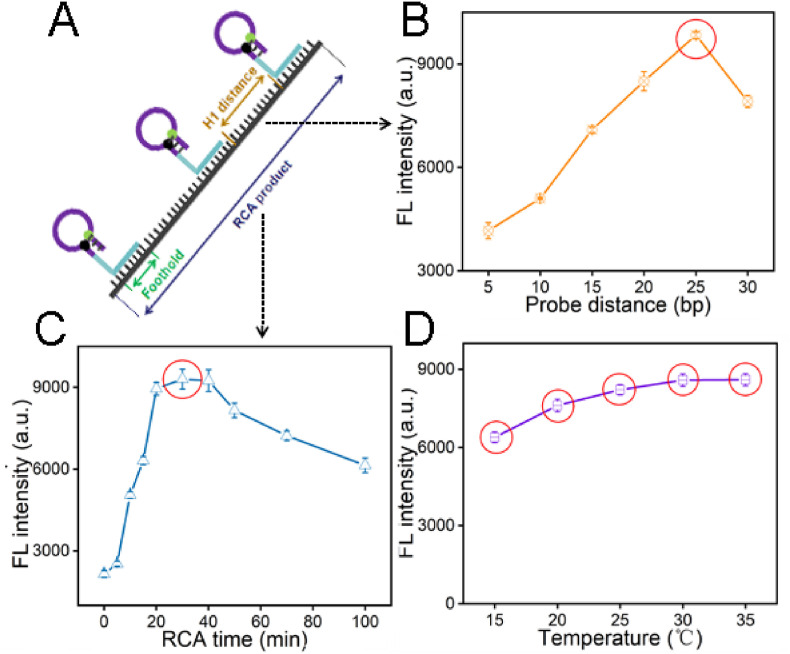
Fig. 2.
Optimize of the experimental parameters of DNHCR. (A) The structure of DNA nanoscaffolds. (B) The effect of different separation distances of H1 probes assembled on long DNA strands. (C) The influence of the RCA time from 0 to 100 min. (D) The effect of the temperature from 15 °C to 35 °C. All the values selected by the red circle are the best condition. The data error bars indicate mean ± SD (n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)