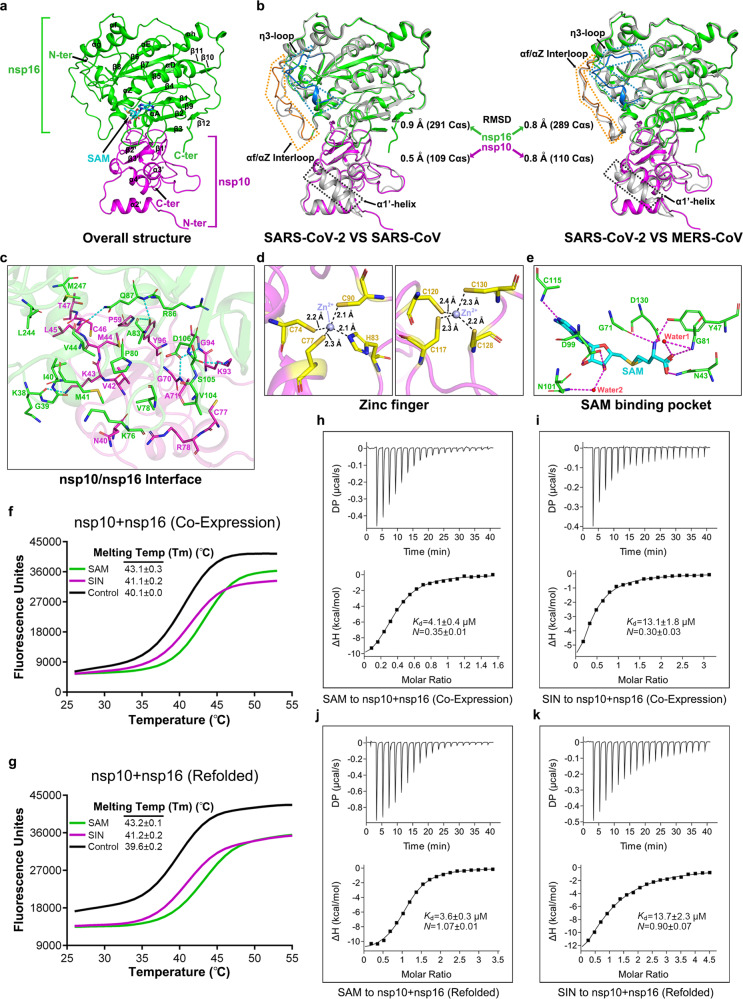
Fig. 1.
Structure of SARS-CoV-2 nsp10/nsp16 hetero-dimer. a Overall structure of the hetero-complex formed between nsp10 (magenta) and nsp16 (green). The secondary structural elements and the bound SAM molecule are labeled. b Superimposition of the nsp10/nsp16 structure of SARS-CoV-2 onto those of SARS-CoV (left panel, PDB: 3R24) and MERS-CoV (right panel, PDB: 5YN5). The color scheme for our structure is the same as in a, and the SARS-CoV and MERS-CoV structures are shown in gray. Those elements exhibiting variant conformations are highlighted and marked. c Detailed interactions at the nsp10/nsp16 binding interface. Dashed lines indicate hydrogen-bonds. d A magnified view on the two zinc-finger motifs in nsp10. e Detailed interactions between SAM and nsp16 at the SAM-binding pocket. f, g Interaction of SAM and SIN with SARS-CoV-2 nsp10/nsp16 characterized by differential scanning fluorimetry (DSF). The DSF data obtained with the protein-complex purified directly from E. coli (designated as co-expression) and the protein-complex after the denaturing-refolding cycle (designated as refolded) are shown in f and g, respectively. h–k Binding of SAM and SIN to SARS-CoV-2 nsp10/nsp16 characterized by isothermal titration calorimetry (ITC). h SAM to nsp10/nsp16 (co-expression). i SIN to nsp10/nsp16 (co-expression). j SAM to nsp10/nsp16 (refolded). k SIN to nsp10/nsp16 (refolded)