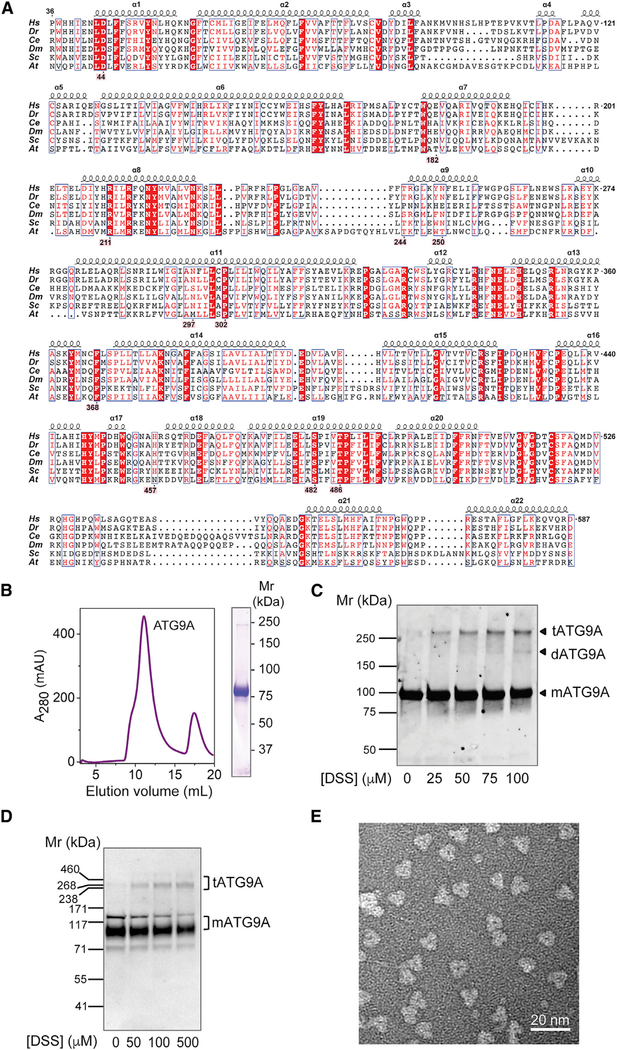
Figure 1. Trimeric Structure of Human ATG9A.
(A) Sequence alignment of the ATG9A core structure. Alignment is numbered according to human ATG9A, comprising amino acids 36–587 (from 839 total). Selected orthologs range in size from 839 to 997 amino acids; depicted are H. sapiens, D. rerio (36–583),C. elegans (102–663), D. melanogaster (87–644), S. cerevisiae (296–843), and A. thaliana (64–620) orthologs. Red boxes highlight 100% sequence identity. Residues highlighted in pink make notable structural contributions. Alignment was generated using the PRALINE webserver, and the figure was adapted from ESPript 3.0 output.
(B) Size-exclusion chromatogram and SDS-PAGE of LMNG-solubilized human ATG9A used for cryo-EM structure determination.
(C) Membranes containing ATG9A-mVenus fusion protein were cross-linked with DSS, run on SDS-PAGE, and imaged with in-gel fluorescence.
(D) Intact HeLa cells were cross-linked with different concentrations of DSS and analyzed by SDS-PAGE and immunoblotting with antibody to endogenous ATG9A. In (C) and (D), the positions of molecular mass markers are indicated on the left, and the positions of ATG9A monomer (mATG9A, Mr ~94 kDa), dimer (dATG9A, Mr ~188 kDa), and trimer (tATG9A, Mr ~282 kDa) are indicated on the right.
(E) Representative negative stain EM image of ATG9A particles, revealing trimeric architecture.